Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T57291
(Former ID: TTDR00363)
|
|||||
| Target Name |
PP2A B subunit isoform R5-alpha (PPP2R5A)
|
|||||
| Synonyms |
Serine/threonine-protein phosphatase 2A 56 kDa regulatory subunit alpha isoform; Protein phosphatase-2A; PR61alpha; PP2A, B subunit, R5 alpha isoform; PP2A, B subunit, PR61 alpha isoform; PP2A, B subunit, B56 alpha isoform; PP2A, B subunit, B' alpha isoform; PP2A B subunit isoform PR61-alpha; PP2A B subunit isoform B56-alpha; PP2A B subunit isoform B'-alpha
Click to Show/Hide
|
|||||
| Gene Name |
PPP2R5A
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
The B regulatory subunit might modulate substrate selectivity and catalytic activity, and also might direct the localization of the catalytic enzyme to a particular subcellular compartment.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MSSSSPPAGAASAAISASEKVDGFTRKSVRKAQRQKRSQGSSQFRSQGSQAELHPLPQLK
DATSNEQQELFCQKLQQCCILFDFMDSVSDLKSKEIKRATLNELVEYVSTNRGVIVESAY SDIVKMISANIFRTLPPSDNPDFDPEEDEPTLEASWPHIQLVYEFFLRFLESPDFQPSIA KRYIDQKFVQQLLELFDSEDPRERDFLKTVLHRIYGKFLGLRAFIRKQINNIFLRFIYET EHFNGVAELLEILGSIINGFALPLKAEHKQFLMKVLIPMHTAKGLALFHAQLAYCVVQFL EKDTTLTEPVIRGLLKFWPKTCSQKEVMFLGEIEEILDVIEPTQFKKIEEPLFKQISKCV SSSHFQVAERALYFWNNEYILSLIEENIDKILPIMFASLYKISKEHWNPTIVALVYNVLK TLMEMNGKLFDDLTSSYKAERQREKKKELEREELWKKLEELKLKKALEKQNSAYNMHSIL SNTSAE Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Methyl L-leucinate | Ligand Info | |||||
| Structure Description | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | PDB:6NTS | ||||
| Method | Electron microscopy | Resolution | 3.63 Å | Mutation | No | [2] |
| PDB Sequence |
QELFCQKLQQ
77 CCILFDFMDS87 VSDLKSKEIK97 RATLNELVEY107 VSTNRGVIVE117 SAYSDIVKMI 127 SANIFRTLPP137 SDNPDFDPEE147 DEPTLEASWP157 HIQLVYEFFL167 RFLESPDFQP 177 SIAKRYIDQK187 FVQQLLELFD197 SEDPRERDFL207 KTVLHRIYGK217 FLGLRAFIRK 227 QINNIFLRFI237 YETEHFNGVA247 ELLEILGSII257 NGFALPLKAE267 HKQFLMKVLI 277 PMHTAKGLAL287 FHAQLAYCVV297 QFLEKDTTLT307 EPVIRGLLKF317 WPKTCSQKEV 327 MFLGEIEEIL337 DVIEPTQFKK347 IEEPLFKQIS357 KCVSSSHFQV367 AERALYFWNN 377 EYILSLIEEN387 IDKILPIMFA397 SLYKISKEHW407 NPTIVALVYN417 VLKTLMEMNG 427 K
|
|||||
|
|
||||||
| Ligand Name: N-[(1R,2R,3S)-2-hydroxy-3-(10H-phenoxazin-10-yl)cyclohexyl]-4-(trifluoromethoxy)benzene-1-sulfonamide | Ligand Info | |||||
| Structure Description | Protein Phosphatase 2A (Aalpha-B56alpha-Calpha) holoenzyme in complex with a Small Molecule Activator of PP2A (SMAP) | PDB:6NTS | ||||
| Method | Electron microscopy | Resolution | 3.63 Å | Mutation | No | [2] |
| PDB Sequence |
QELFCQKLQQ
77 CCILFDFMDS87 VSDLKSKEIK97 RATLNELVEY107 VSTNRGVIVE117 SAYSDIVKMI 127 SANIFRTLPP137 SDNPDFDPEE147 DEPTLEASWP157 HIQLVYEFFL167 RFLESPDFQP 177 SIAKRYIDQK187 FVQQLLELFD197 SEDPRERDFL207 KTVLHRIYGK217 FLGLRAFIRK 227 QINNIFLRFI237 YETEHFNGVA247 ELLEILGSII257 NGFALPLKAE267 HKQFLMKVLI 277 PMHTAKGLAL287 FHAQLAYCVV297 QFLEKDTTLT307 EPVIRGLLKF317 WPKTCSQKEV 327 MFLGEIEEIL337 DVIEPTQFKK347 IEEPLFKQIS357 KCVSSSHFQV367 AERALYFWNN 377 EYILSLIEEN387 IDKILPIMFA397 SLYKISKEHW407 NPTIVALVYN417 VLKTLMEMNG 427 K
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| mRNA surveillance pathway | hsa03015 | Affiliated Target |
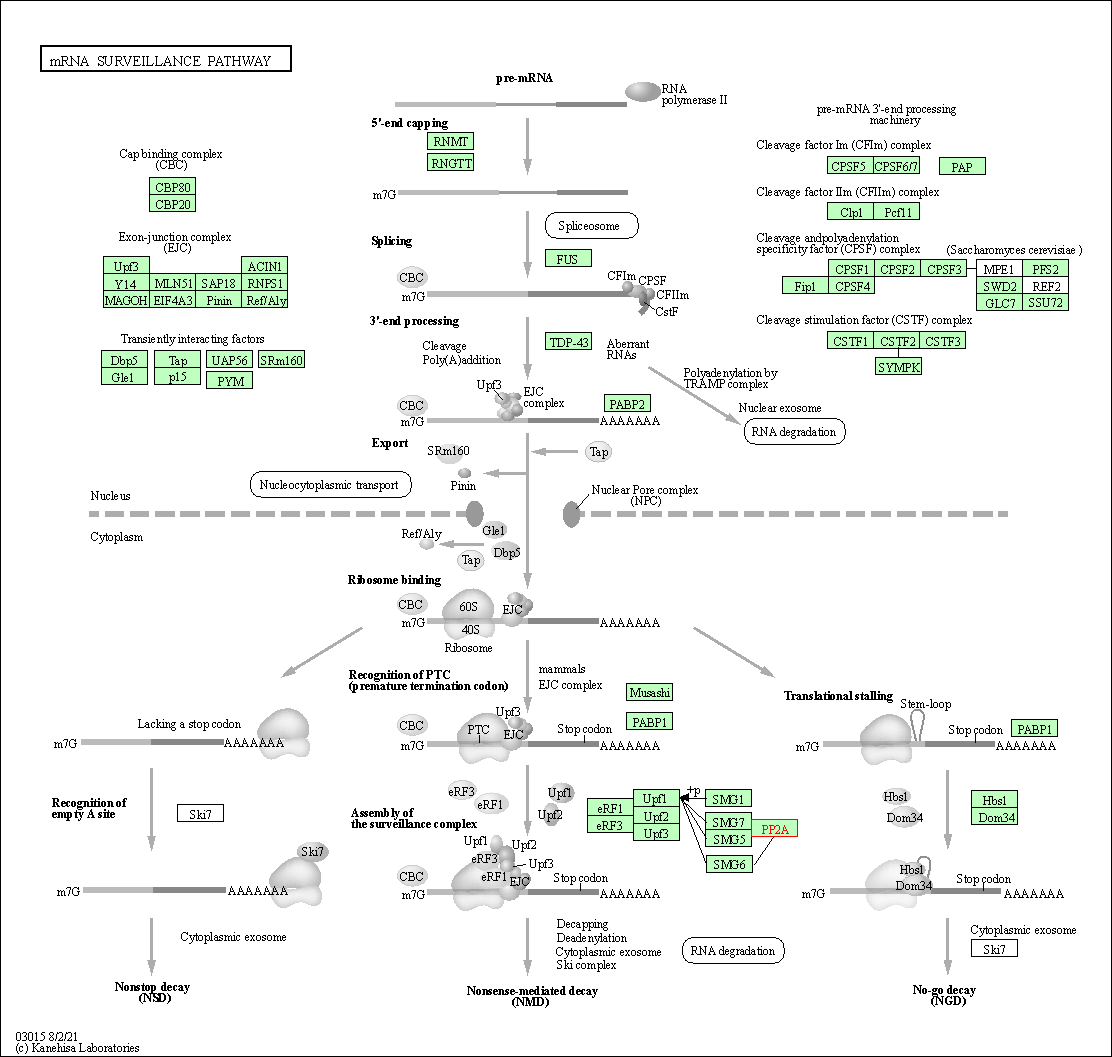
|
| Class: Genetic Information Processing => Translation | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |
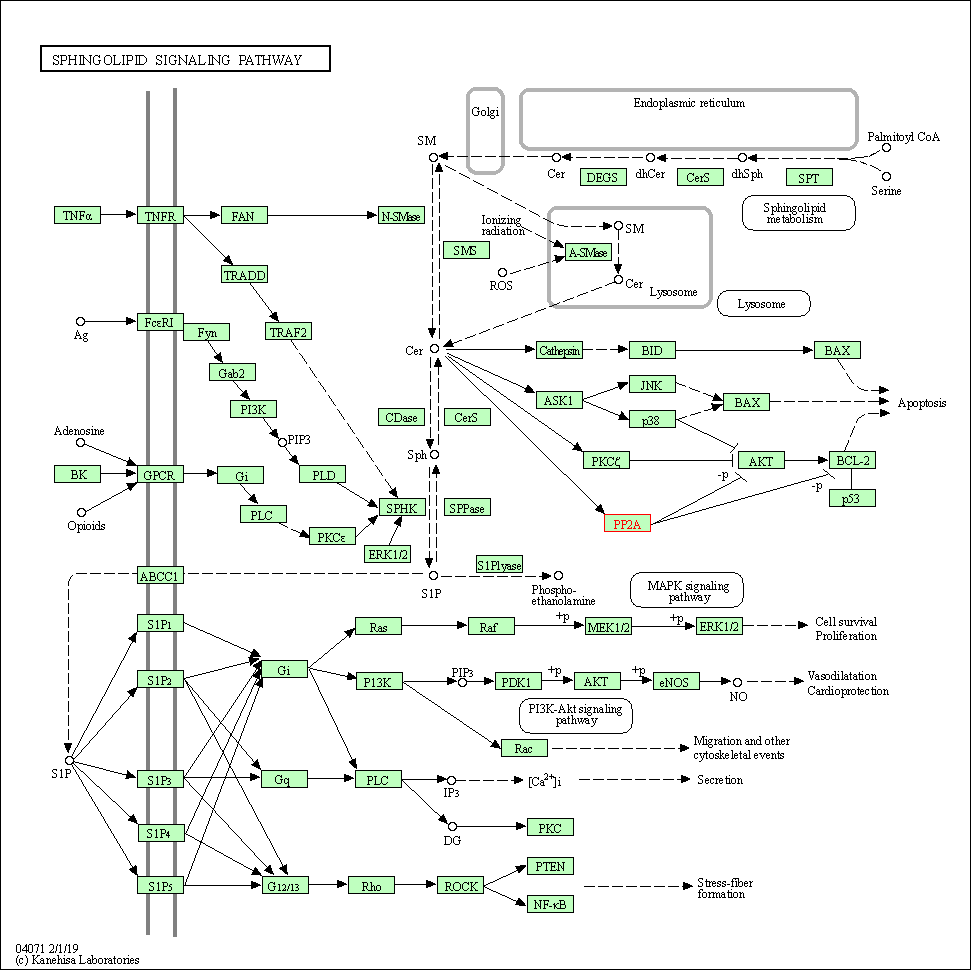
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |
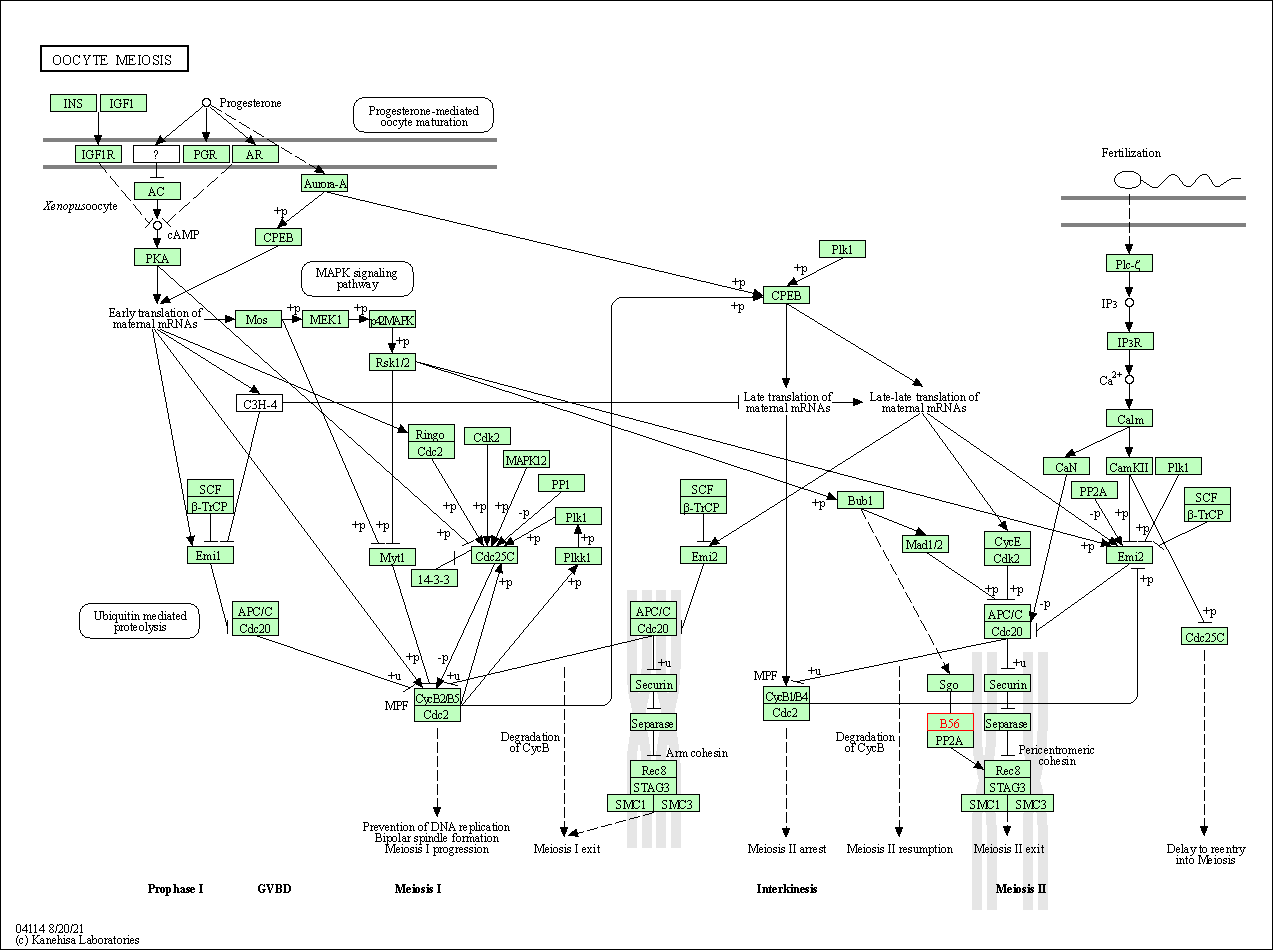
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
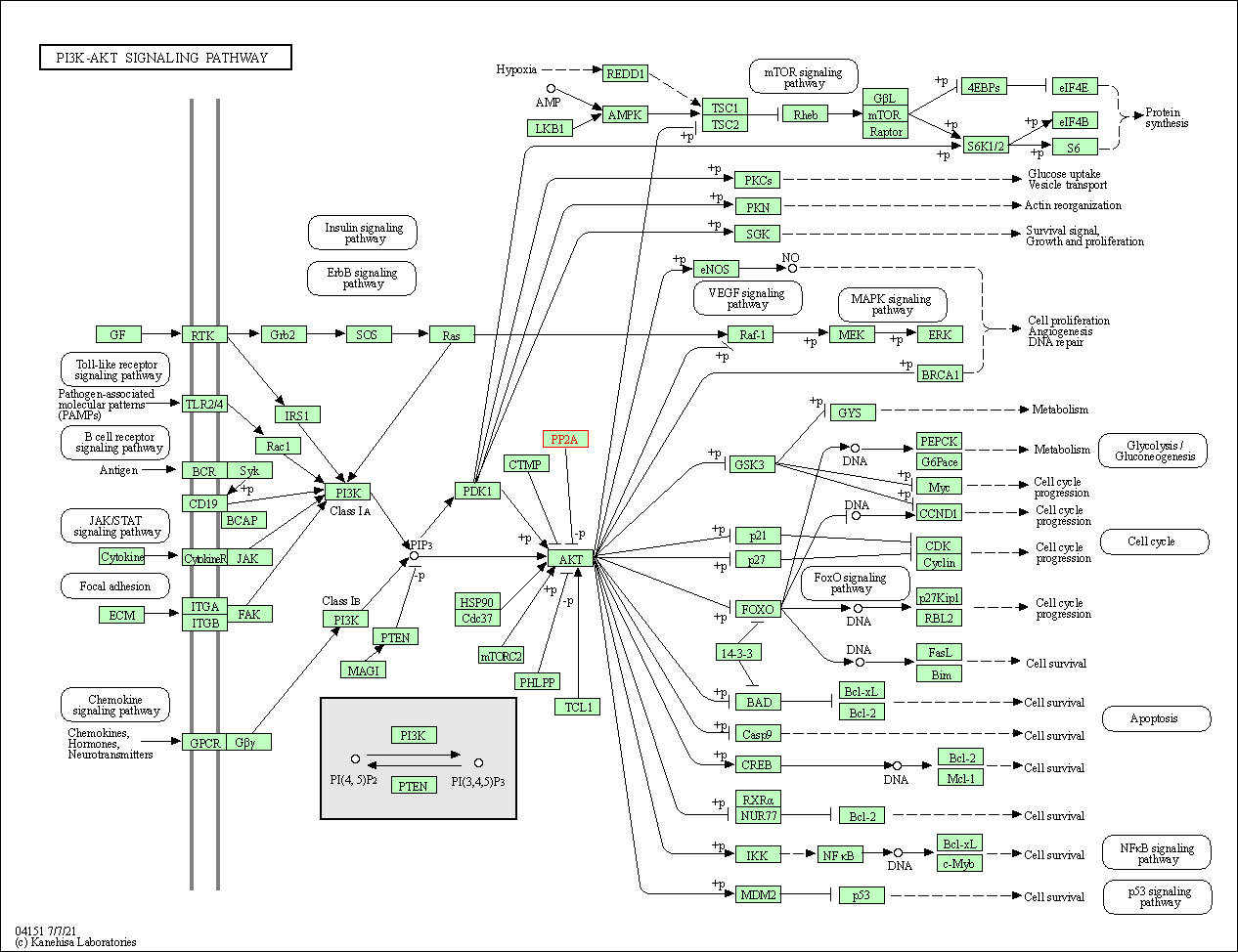
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| AMPK signaling pathway | hsa04152 | Affiliated Target |
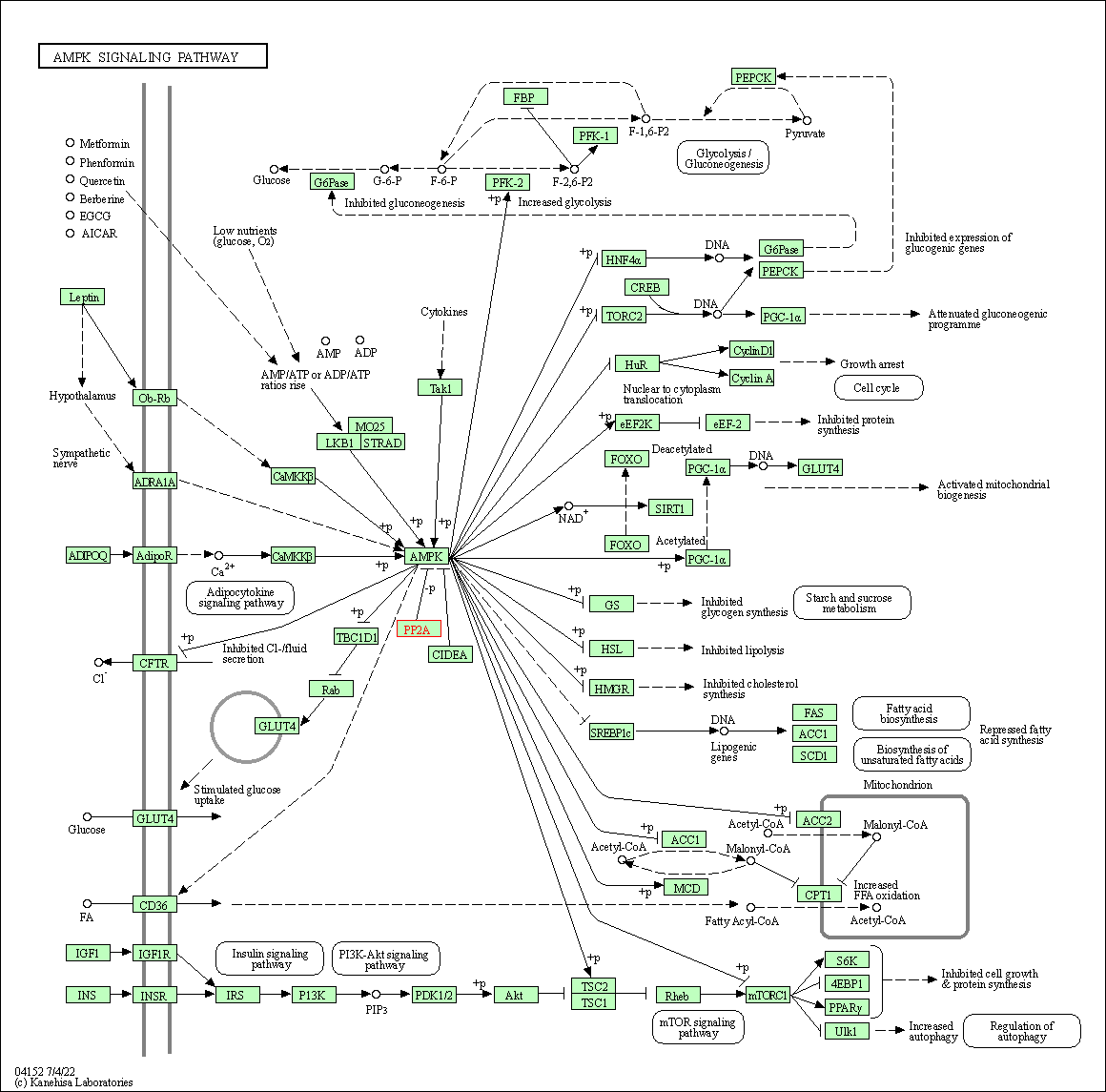
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |
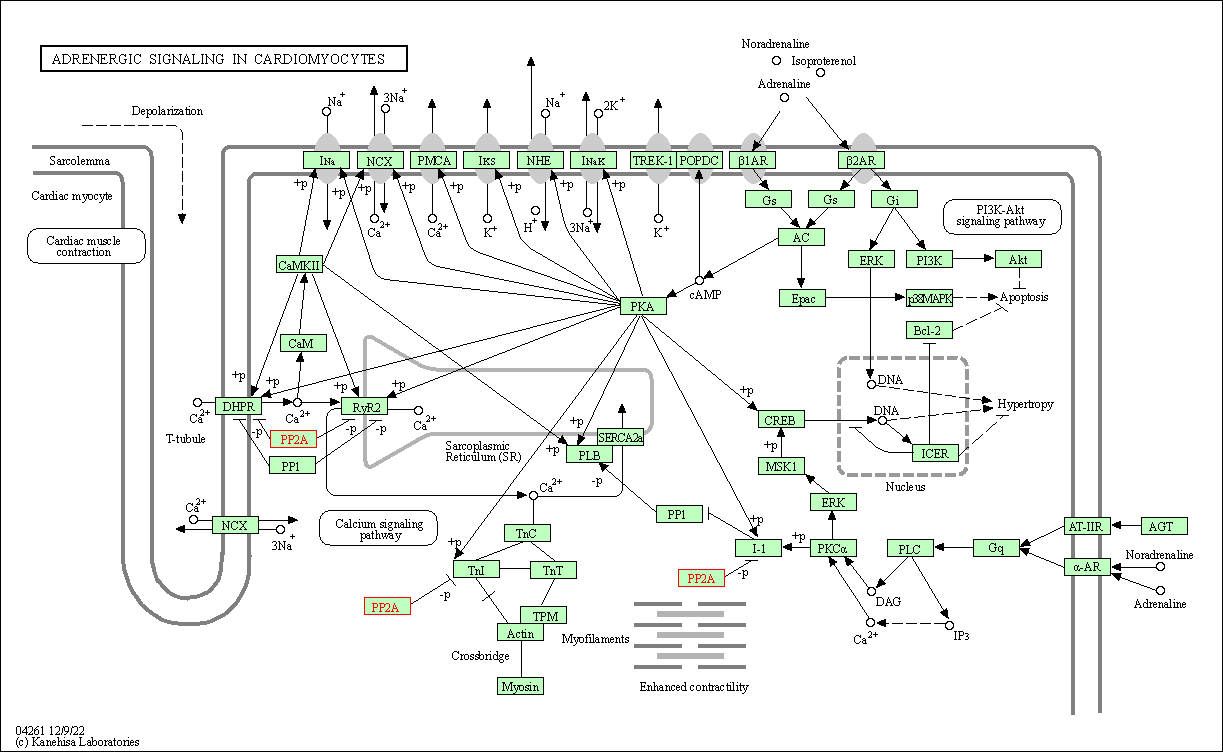
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
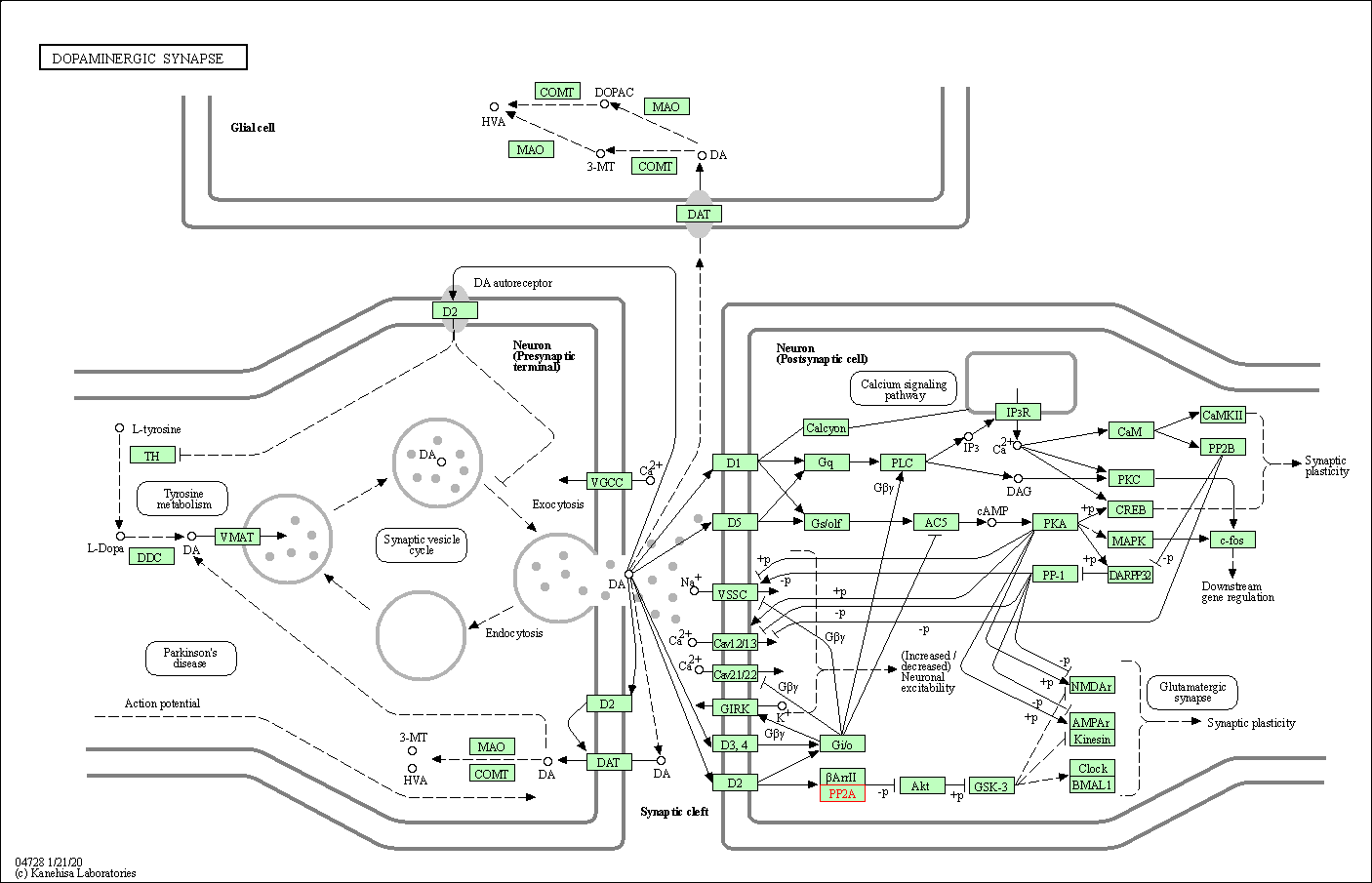
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 15 | Degree centrality | 1.61E-03 | Betweenness centrality | 7.68E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 2.43E-01 | Radiality | 1.43E+01 | Clustering coefficient | 3.62E-01 |
| Neighborhood connectivity | 5.33E+01 | Topological coefficient | 1.04E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Structure-activity relationship of cantharidin derivatives to protein phosphatases 1, 2A1, and 2B, Bioorg. Med. Chem. Lett. 7(14):1833-1836 (1997). | |||||
| REF 2 | Selective PP2A Enhancement through Biased Heterotrimer Stabilization. Cell. 2020 Apr 30;181(3):688-701.e16. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

