Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T47721
(Former ID: TTDR00535)
|
|||||
| Target Name |
Intrinsic factor-B12 receptor (CUBN)
|
|||||
| Synonyms |
Intrinsic factor-vitamin B12 receptor; Cubilin; CUBN
Click to Show/Hide
|
|||||
| Gene Name |
CUBN
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Cotransporter which plays a role in lipoprotein, vitamin and iron metabolism, by facilitating their uptake. Binds to ALB, MB, Kappa and lambda-light chains, TF, hemoglobin, GC, SCGB1A1, APOA1, high density lipoprotein, and the GIF-cobalamin complex. The binding of all ligands requires calcium. Serves as important transporter in several absorptive epithelia, including intestine, renal proximal tubules and embryonic yolk sac. Interaction with LRP2 mediates its trafficking throughout vesicles and facilitates the uptake of specific ligands like GC, hemoglobin, ALB, TF and SCGB1A1. Interaction with AMN controls its trafficking to the plasma membrane and facilitates endocytosis of ligands. May play an important role in the development of the peri-implantation embryo through internalization of APOA1 and cholesterol. Binds to LGALS3 at the maternal-fetal interface.
Click to Show/Hide
|
|||||
| BioChemical Class |
Growth factor
|
|||||
| UniProt ID | ||||||
| Sequence |
MMNMSLPFLWSLLTLLIFAEVNGEAGELELQRQKRSINLQQPRMATERGNLVFLTGSAQN
IEFRTGSLGKIKLNDEDLSECLHQIQKNKEDIIELKGSAIGLPQNISSQIYQLNSKLVDL ERKFQGLQQTVDKKVCSSNPCQNGGTCLNLHDSFFCICPPQWKGPLCSADVNECEIYSGT PLSCQNGGTCVNTMGSYSCHCPPETYGPQCASKYDDCEGGSVARCVHGICEDLMREQAGE PKYSCVCDAGWMFSPNSPACTLDRDECSFQPGPCSTLVQCFNTQGSFYCGACPTGWQGNG YICEDINECEINNGGCSVAPPVECVNTPGSSHCQACPPGYQGDGRVCTLTDICSVSNGGC HPDASCSSTLGSLPLCTCLPGYTGNGYGPNGCVQLSNICLSHPCLNGQCIDTVSGYFCKC DSGWTGVNCTENINECLSNPCLNGGTCVDGVDSFSCECTRLWTGALCQVPQQVCGESLSG INGSFSYRSPDVGYVHDVNCFWVIKTEMGKVLRITFTFFRLESMDNCPHEFLQVYDGDSS SAFQLGRFCGSSLPHELLSSDNALYFHLYSEHLRNGRGFTVRWETQQPECGGILTGPYGS IKSPGYPGNYPPGRDCVWIVVTSPDLLVTFTFGTLSLEHHDDCNKDYLEIRDGPLYQDPL LGKFCTTFSVPPLQTTGPFARIHFHSDSQISDQGFHITYLTSPSDLRCGGNYTDPEGELF LPELSGPFTHTRQCVYMMKQPQGEQIQINFTHVELQCQSDSSQNYIEVRDGETLLGKVCG NGTISHIKSITNSVWIRFKIDASVEKASFRAVYQVACGDELTGEGVIRSPFFPNVYPGER TCRWTIHQPQSQVILLNFTVFEIGSSAHCETDYVEIGSSSILGSPENKKYCGTDIPSFIT SVYNFLYVTFVKSSSTENHGFMAKFSAEDLACGEILTESTGTIQSPGHPNVYPHGINCTW HILVQPNHLIHLMFETFHLEFHYNCTNDYLEVYDTDSETSLGRYCGKSIPPSLTSSGNSL MLVFVTDSDLAYEGFLINYEAISAATACLQDYTDDLGTFTSPNFPNNYPNNWECIYRITV RTGQLIAVHFTNFSLEEAIGNYYTDFLEIRDGGYEKSPLLGIFYGSNLPPTIISHSNKLW LKFKSDQIDTRSGFSAYWDGSSTGCGGNLTTSSGTFISPNYPMPYYHSSECYWWLKSSHG SAFELEFKDFHLEHHPNCTLDYLAVYDGPSSNSHLLTQLCGDEKPPLIRSSGDSMFIKLR TDEGQQGRGFKAEYRQTCENVVIVNQTYGILESIGYPNPYSENQHCNWTIRATTGNTVNY TFLAFDLEHHINCSTDYLELYDGPRQMGRYCGVDLPPPGSTTSSKLQVLLLTDGVGRREK GFQMQWFVYGCGGELSGATGSFSSPGFPNRYPPNKECIWYIRTDPGSSIQLTIHDFDVEY HSRCNFDVLEIYGGPDFHSPRIAQLCTQRSPENPMQVSSTGNELAIRFKTDLSINGRGFN ASWQAVTGGCGGIFQAPSGEIHSPNYPSPYRSNTDCSWVIRVDRNHRVLLNFTDFDLEPQ DSCIMAYDGLSSTMSRLARTCGREQLANPIVSSGNSLFLRFQSGPSRQNRGFRAQFRQAC GGHILTSSFDTVSSPRFPANYPNNQNCSWIIQAQPPLNHITLSFTHFELERSTTCARDFV EILDGGHEDAPLRGRYCGTDMPHPITSFSSALTLRFVSDSSISAGGFHTTVTASVSACGG TFYMAEGIFNSPGYPDIYPPNVECVWNIVSSPGNRLQLSFISFQLEDSQDCSRDFVEIRE GNATGHLVGRYCGNSFPLNYSSIVGHTLWVRFISDGSGSGTGFQATFMKIFGNDNIVGTH GKVASPFWPENYPHNSNYQWTVNVNASHVVHGRILEMDIEEIQNCYYDKLRIYDGPSIHA RLIGAYCGTQTESFSSTGNSLTFHFYSDSSISGKGFLLEWFAVDAPDGVLPTIAPGACGG FLRTGDAPVFLFSPGWPDSYSNRVDCTWLIQAPDSTVELNILSLDIESHRTCAYDSLVIR DGDNNLAQQLAVLCGREIPGPIRSTGEYMFIRFTSDSSVTRAGFNASFHKSCGGYLHADR GIITSPKYPETYPSNLNCSWHVLVQSGLTIAVHFEQPFQIPNGDSSCNQGDYLVLRNGPD ICSPPLGPPGGNGHFCGSHASSTLFTSDNQMFVQFISDHSNEGQGFKIKYEAKSLACGGN VYIHDADSAGYVTSPNHPHNYPPHADCIWILAAPPETRIQLQFEDRFDIEVTPNCTSNYL ELRDGVDSDAPILSKFCGTSLPSSQWSSGEVMYLRFRSDNSPTHVGFKAKYSIAQCGGRV PGQSGVVESIGHPTLPYRDNLFCEWHLQGLSGHYLTISFEDFNLQNSSGCEKDFVEIWDN HTSGNILGRYCGNTIPDSIDTSSNTAVVRFVTDGSVTASGFRLRFESSMEECGGDLQGSI GTFTSPNYPNPNPHGRICEWRITAPEGRRITLMFNNLRLATHPSCNNEHVIVFNGIRSNS PQLEKLCSSVNVSNEIKSSGNTMKVIFFTDGSRPYGGFTASYTSSEDAVCGGSLPNTPEG NFTSPGYDGVRNYSRNLNCEWTLSNPNQGNSSISIHFEDFYLESHQDCQFDVLEFRVGDA DGPLMWRLCGPSKPTLPLVIPYSQVWIHFVTNERVEHIGFHAKYSFTDCGGIQIGDSGVI TSPNYPNAYDSLTHCSSLLEAPQGHTITLTFSDFDIEPHTTCAWDSVTVRNGGSPESPII GQYCGNSNPRTIQSGSNQLVVTFNSDHSLQGGGFYATWNTQTLGCGGIFHSDNGTIRSPH WPQNFPENSRCSWTAITHKSKHLEISFDNNFLIPSGDGQCQNSFVKVWAGTEEVDKALLA TGCGNVAPGPVITPSNTFTAVFQSQEAPAQGFSASFVSRCGSNFTGPSGYIISPNYPKQY DNNMNCTYVIEANPLSVVLLTFVSFHLEARSAVTGSCVNDGVHIIRGYSVMSTPFATVCG DEMPAPLTIAGPVLLNFYSNEQITDFGFKFSYRIISCGGVFNFSSGIITSPAYSYADYPN DMHCLYTITVSDDKVIELKFSDFDVVPSTSCSHDYLAIYDGANTSDPLLGKFCGSKRPPN VKSSNNSMLLVFKTDSFQTAKGWKMSFRQTLGPQQGCGGYLTGSNNTFASPDSDSNGMYD KNLNCVWIIIAPVNKVIHLTFNTFALEAASTRQRCLYDYVKLYDGDSENANLAGTFCGST VPAPFISSGNFLTVQFISDLTLEREGFNATYTIMDMPCGGTYNATWTPQNISSPNSSDPD VPFSICTWVIDSPPHQQVKITVWALQLTSQDCTQNYLQLQDSPQGHGNSRFQFCGRNASA VPVFYSSMSTAMVIFKSGVVNRNSRMSFTYQIADCNRDYHKAFGNLRSPGWPDNYDNDKD CTVTLTAPQNHTISLFFHSLGIENSVECRNDFLEVRNGSNSNSPLLGKYCGTLLPNPVFS QNNELYLRFKSDSVTSDRGYEIIWTSSPSGCGGTLYGDRGSFTSPGYPGTYPNNTYCEWV LVAPAGRLVTINFYFISIDDPGDCVQNYLTLYDGPNASSPSSGPYCGGDTSIAPFVASSN QVFIKFHADYARRPSAFRLTWDS Click to Show/Hide
|
|||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Vitamin digestion and absorption | hsa04977 | Affiliated Target |
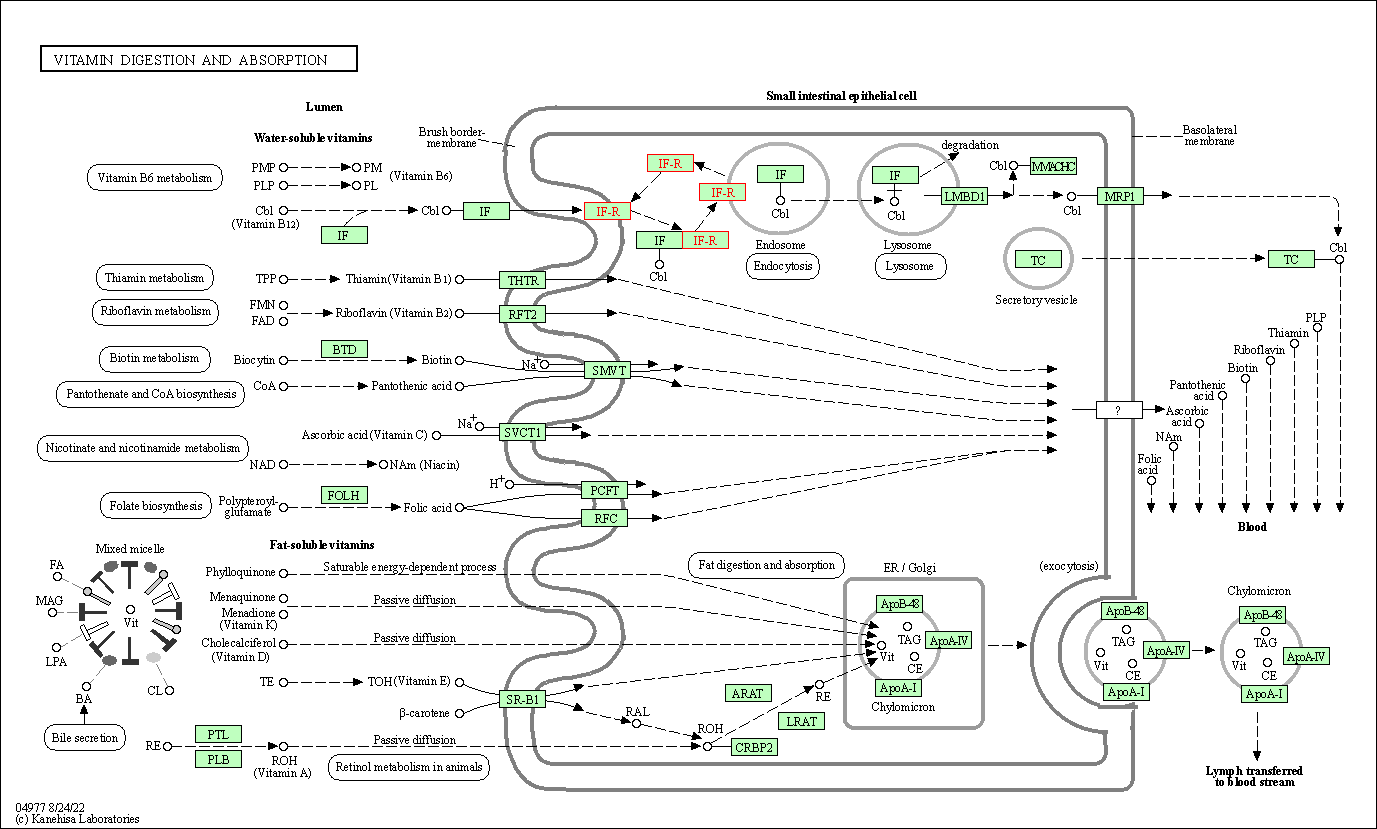
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 2.26E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.88E-01 | Radiality | 1.32E+01 | Clustering coefficient | 3.33E-01 |
| Neighborhood connectivity | 1.38E+01 | Topological coefficient | 2.10E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Sodium reabsorption in aldosterone-sensitive distal nephron: news and contributions from genetically engineered animals. Curr Opin Nephrol Hypertens. 2001 Jan;10(1):39-47. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

