Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T46263
(Former ID: TTDI02011)
|
|||||
| Target Name |
Lymphocyte activation antigen 4F2 (SLC3A2)
|
|||||
| Synonyms |
SLC3A2; Lymphocyte activation antigen 4F2 large subunit; CD98; 4F2hc; 4F2 heavy chain antigen; 4F2 cellsurface antigen heavy chain
Click to Show/Hide
|
|||||
| Gene Name |
SLC3A2
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Acute myeloid leukaemia [ICD-11: 2A60] | |||||
| Function |
Required for the function of light chain amino-acid transporters. Involved in sodium-independent, high-affinity transport of large neutral amino acids such as phenylalanine, tyrosine, leucine, arginine and tryptophan. Involved in guiding and targeting of LAT1 and LAT2 to the plasma membrane. When associated with SLC7A6 or SLC7A7 acts as an arginine/glutamine exchanger, following an antiport mechanism for amino acid transport, influencing arginine release in exchange for extracellular amino acids. Plays a role in nitric oxide synthesis in human umbilical vein endothelial cells (HUVECs) via transport of L-arginine. Required for normal and neoplastic cell growth. When associated with SLC7A5/LAT1, is also involved in the transport of L-DOPA across the blood-brain barrier, and that of thyroid hormones triiodothyronine (T3) and thyroxine (T4) across the cell membrane in tissues such as placenta. Involved in the uptake of methylmercury (MeHg) when administered as the L-cysteine or D,L-homocysteine complexes, and hence plays a role in metal ion homeostasis and toxicity. When associated with SLC7A5 or SLC7A8, involved in the cellular activity of small molecular weight nitrosothiols, via the stereoselective transport of L- nitrosocysteine (L-CNSO) across the transmembrane. Together with ICAM1, regulates the transport activity LAT2 in polarized intestinal cells, by generating and delivering intracellular signals. Whenassociated with SLC7A5, plays an important role in transporting L-leucine from the circulating blood to the retina across the inner blood-retinal barrier.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MELQPPEASIAVVSIPRQLPGSHSEAGVQGLSAGDDSELGSHCVAQTGLELLASGDPLPS
ASQNAEMIETGSDCVTQAGLQLLASSDPPALASKNAEVTGTMSQDTEVDMKEVELNELEP EKQPMNAASGAAMSLAGAEKNGLVKIKVAEDEAEAAAAAKFTGLSKEELLKVAGSPGWVR TRWALLLLFWLGWLGMLAGAVVIIVRAPRCRELPAQKWWHTGALYRIGDLQAFQGHGAGN LAGLKGRLDYLSSLKVKGLVLGPIHKNQKDDVAQTDLLQIDPNFGSKEDFDSLLQSAKKK SIRVILDLTPNYRGENSWFSTQVDTVATKVKDALEFWLQAGVDGFQVRDIENLKDASSFL AEWQNITKGFSEDRLLIAGTNSSDLQQILSLLESNKDLLLTSSYLSDSGSTGEHTKSLVT QYLNATGNRWCSWSLSQARLLTSFLPAQLLRLYQLMLFTLPGTPVFSYGDEIGLDAAALP GQPMEAPVMLWDESSFPDIPGAVSANMTVKGQSEDPGSLLSLFRRLSDQRSKERSLLHGD FHAFSAGPGLFSYIRHWDQNERFLVVLNFGDVGLSAGLQASDLPASASLPAKADLLLSTQ PGREEGSPLELERLKLEPHEGLLLRFPYAA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T56RFU | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 1 Clinical Trial Drugs | + | ||||
| 1 | IGN523 | Drug Info | Phase 1 | Acute myeloid leukaemia | [2], [3] | |
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Cholesterol | Ligand Info | |||||
| Structure Description | LAT1-CD98hc complex bound to MEM-108 Fab | PDB:6JMQ | ||||
| Method | Electron microscopy | Resolution | 3.31 Å | Mutation | No | [4] |
| PDB Sequence |
RTRWALLLLF
189 WLGWLGMLAG199 AVVIIVRAPR209 CRELPAQKWW219 HTGALYRIGD229 LQAFQGHGAG 239 NLAGLKGRLD249 YLSSLKVKGL259 VLGPIHKNQK269 DDVAQTDLLQ279 IDPNFGSKED 289 FDSLLQSAKK299 KSIRVILDLT309 PNYRGENSWF319 STQVDTVATK329 VKDALEFWLQ 339 AGVDGFQVRD349 IENLKDASSF359 LAEWQNITKG369 FSEDRLLIAG379 TNSSDLQQIL 389 SLLESNKDLL399 LTSSYLSDSG409 STGEHTKSLV419 TQYLNATGNR429 WCSWSLSQAR 439 LLTSFLPAQL449 LRLYQLMLFT459 LPGTPVFSYG469 DEIGLDAAAL479 PGQPMEAPVM 489 LWDESSFPDI499 PGAVSANMTV509 KGQSEDPGSL519 LSLFRRLSDQ529 RSKERSLLHG 539 DFHAFSAGPG549 LFSYIRHWDQ559 NERFLVVLNF569 GDVGLSAGLQ579 ASDLPASASL 589 PAKADLLLST599 QPGREEGSPL609 ELERLKLEPH619 EGLLLRFPYA629 A |
|||||
|
|
||||||
| Ligand Name: Cholesterol hemisuccinate | Ligand Info | |||||
| Structure Description | Overall structure of the LAT1-4F2hc bound with JX-075 | PDB:7DSK | ||||
| Method | Electron microscopy | Resolution | 2.90 Å | Mutation | No | [5] |
| PDB Sequence |
KFTGLSKEEL
170 LKVAGSPGWV180 RTRWALLLLF190 WLGWLGMLAG200 AVVIIVRAPR210 CRELPAQKWW 220 HTGALYRIGD230 LQAFQGHGAG240 NLAGLKGRLD250 YLSSLKVKGL260 VLGPIHKNQK 270 DDVAQTDLLQ280 IDPNFGSKED290 FDSLLQSAKK300 KSIRVILDLT310 PNYRGENSWF 320 STQVDTVATK330 VKDALEFWLQ340 AGVDGFQVRD350 IENLKDASSF360 LAEWQNITKG 370 FSEDRLLIAG380 TNSSDLQQIL390 SLLESNKDLL400 LTSSYLSDSG410 STGEHTKSLV 420 TQYLNATGNR430 WCSWSLSQAR440 LLTSFLPAQL450 LRLYQLMLFT460 LPGTPVFSYG 470 DEIGLDAAAL480 PGQPMEAPVM490 LWDESSFPDI500 PGAVSANMTV510 KGQSEDPGSL 520 LSLFRRLSDQ530 RSKERSLLHG540 DFHAFSAGPG550 LFSYIRHWDQ560 NERFLVVLNF 570 GDVGLSAGLQ580 ASDLPASASL590 PAKADLLLST600 QPGREEGSPL610 ELERLKLEPH 620 EGLLLRFPYA630 A
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| mTOR signaling pathway | hsa04150 | Affiliated Target |
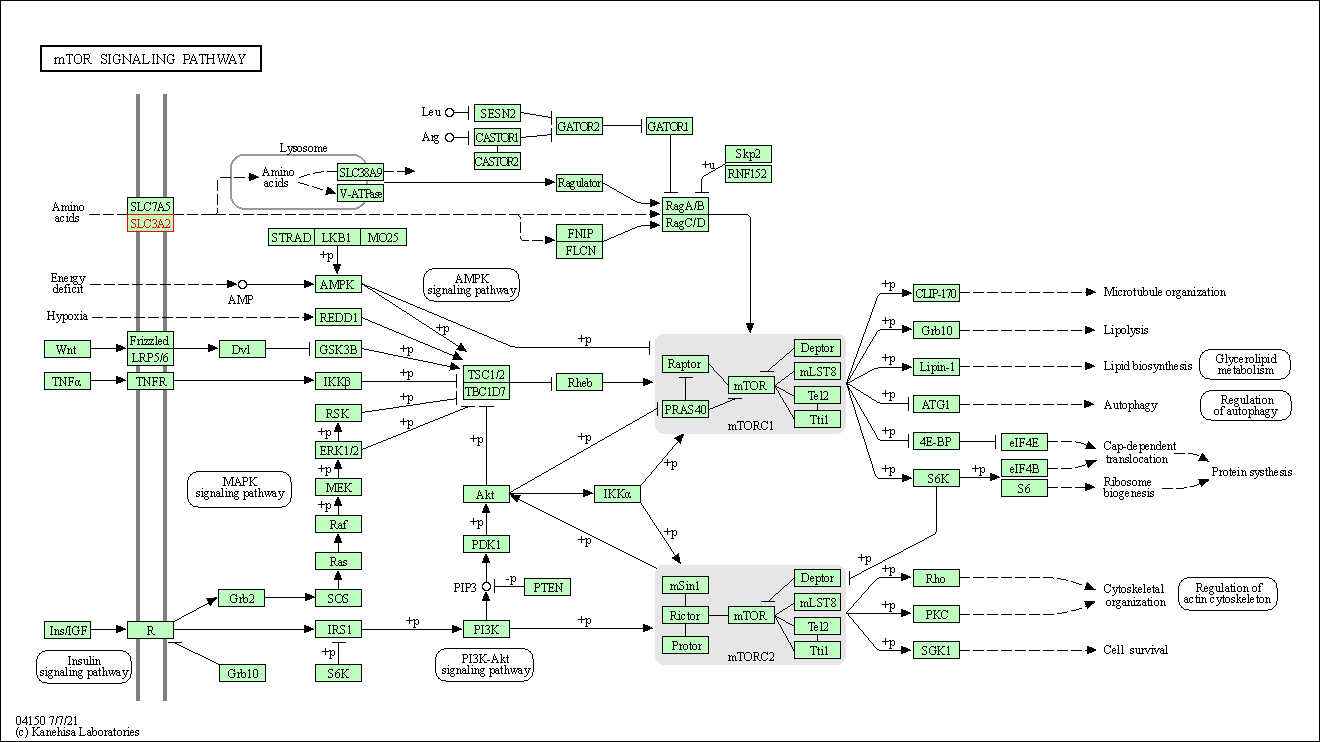
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ferroptosis | hsa04216 | Affiliated Target |
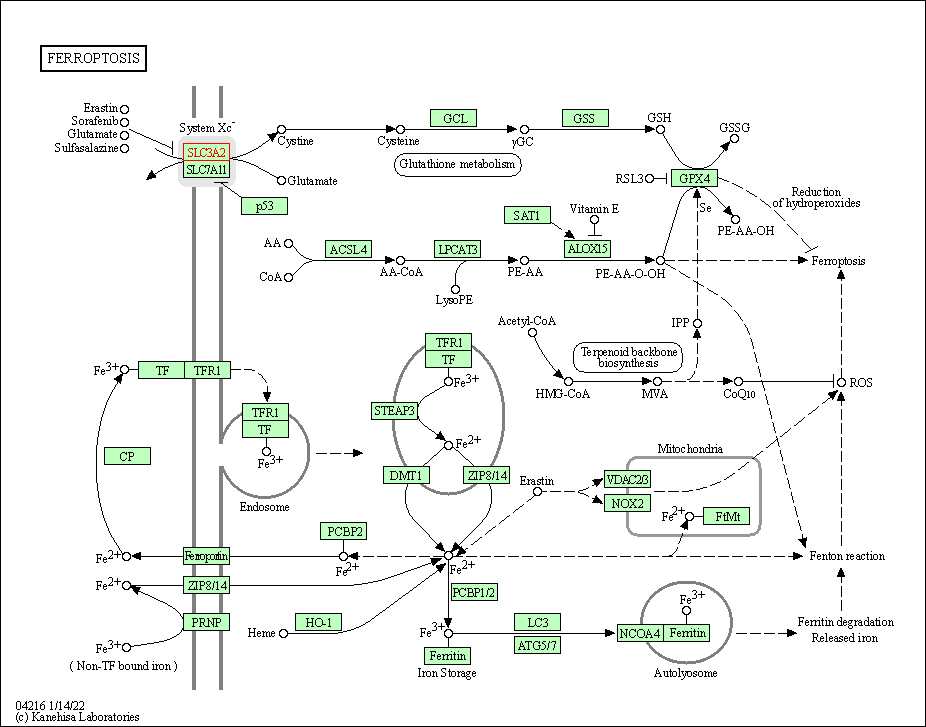
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Protein digestion and absorption | hsa04974 | Affiliated Target |
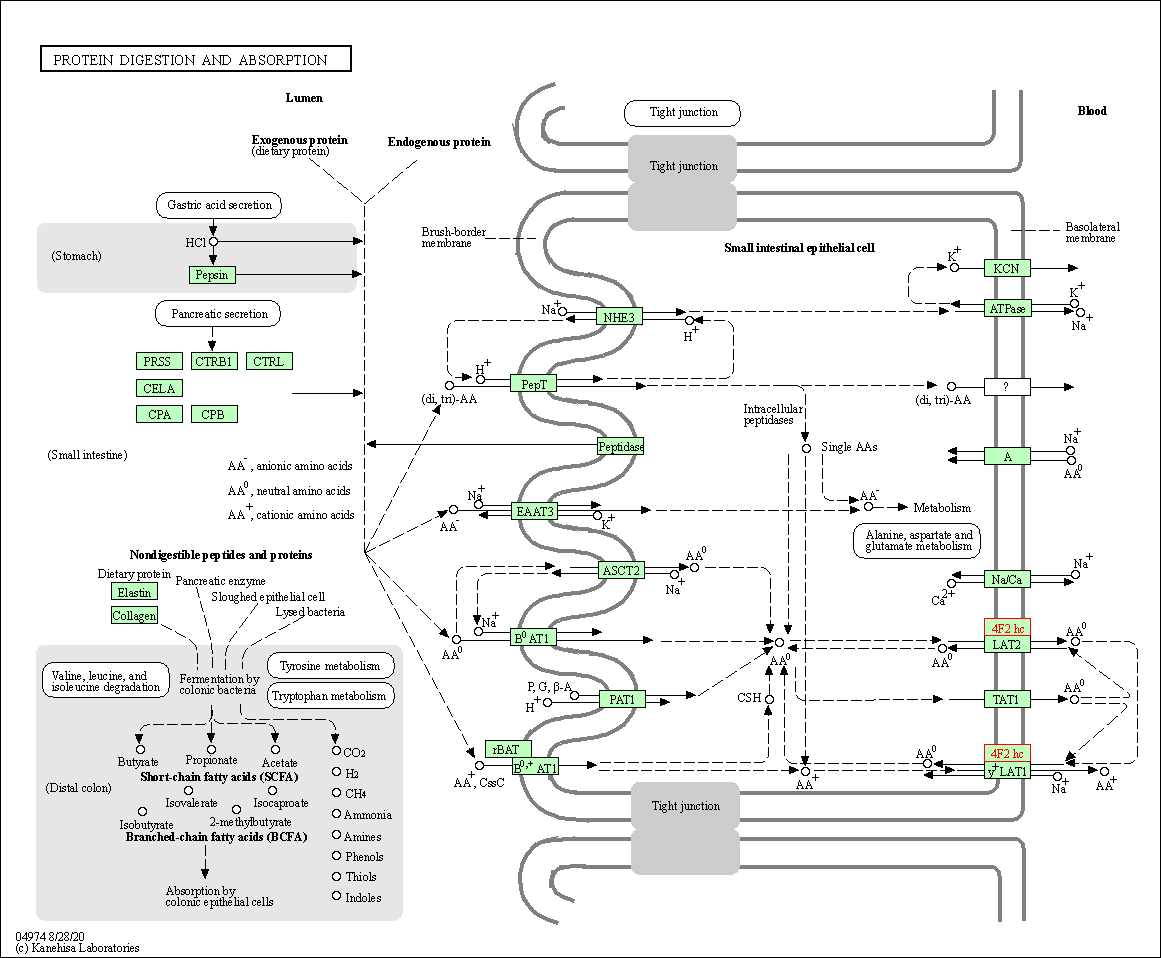
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Degree | 8 | Degree centrality | 8.59E-04 | Betweenness centrality | 1.21E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 1.79E-01 | Radiality | 1.29E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 2.50E+00 | Topological coefficient | 1.36E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 1 KEGG Pathways | + | ||||
| 1 | Protein digestion and absorption | |||||
| NetPath Pathway | [+] 1 NetPath Pathways | + | ||||
| 1 | TCR Signaling Pathway | |||||
| Pathwhiz Pathway | [+] 1 Pathwhiz Pathways | + | ||||
| 1 | Kidney Function | |||||
| PID Pathway | [+] 1 PID Pathways | + | ||||
| 1 | Calcineurin-regulated NFAT-dependent transcription in lymphocytes | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Basigin interactions | |||||
| 2 | Amino acid transport across the plasma membrane | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Transport of inorganic cations/anions and amino acids/oligopeptides | |||||
| 2 | Cell surface interactions at the vascular wall | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Antitumor activity of an anti-CD98 antibody. Int J Cancer. 2015 Aug 1;137(3):710-20. | |||||
| REF 2 | ClinicalTrials.gov (NCT02040506) A Phase I Study of IGN523 in Subjects With Relapsed or Refractory AML. U.S. National Institutes of Health. | |||||
| REF 3 | Clinical pipeline report, company report or official report of the Pharmaceutical Research and Manufacturers of America (PhRMA) | |||||
| REF 4 | Cryo-EM structure of the human L-type amino acid transporter 1 in complex with glycoprotein CD98hc. Nat Struct Mol Biol. 2019 Jun;26(6):510-517. | |||||
| REF 5 | Mechanism of substrate transport and inhibition of the human LAT1-4F2hc amino acid transporter. Cell Discov. 2021 Mar 23;7(1):16. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

