Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T44658
(Former ID: TTDR01281)
|
|||||
| Target Name |
CCCH tandem zinc finger protein ZFP36L1 (ERF-1)
|
|||||
| Synonyms |
mRNA decay activator protein ZFP36L1; Zinc finger protein 36, C3H1 type-like 1; ZFP36-like 1; TPA-induced sequence 11b; ERF-1; EGF-response factor 1; Butyrate response factor 1; BERG36
Click to Show/Hide
|
|||||
| Gene Name |
ZFP36L1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Zinc-finger RNA-binding protein that destabilizes several cytoplasmic AU-rich element (ARE)-containing mRNA transcripts by promoting their poly(A) tail removal or deadenylation, and hence provide a mechanism for attenuating protein synthesis. Acts as a 3'-untranslated region (UTR) ARE mRNA-binding adapter protein to communicate signaling events to the mRNA decay machinery. Functions by recruiting the CCR4-NOT deadenylase complex and components of the cytoplasmic RNA decay machinery to the bound ARE-containing mRNAs, and hence promotes ARE-mediated mRNA deadenylation and decay processes. Induces also the degradation of ARE-containing mRNAs even in absence of poly(A) tail (By similarity). Binds to 3'-UTR ARE of numerous mRNAs. Positively regulates early adipogenesis by promoting ARE-mediated mRNA decay of immediate early genes (IEGs) (By similarity). Promotes ARE-mediated mRNA decay of mineralocorticoid receptor NR3C2 mRNA in response to hypertonic stress. Negatively regulates hematopoietic/erythroid cell differentiation by promoting ARE-mediated mRNA decay of the transcription factor STAT5B mRNA. Positively regulates monocyte/macrophage cell differentiation by promoting ARE-mediated mRNA decay of the cyclin-dependent kinase CDK6 mRNA. Promotes degradation of ARE-containing pluripotency-associated mRNAs in embryonic stem cells (ESCs), such as NANOG, through a fibroblast growth factor (FGF)-induced MAPK-dependent signaling pathway, and hence attenuates ESC self-renewal and positively regulates mesendoderm differentiation (By similarity). May play a role in mediating pro-apoptotic effects in malignant B-cells by promoting ARE-mediated mRNA decay of BCL2 mRNA. In association with ZFP36L2 maintains quiescence on developing B lymphocytes by promoting ARE-mediated decay of several mRNAs encoding cell cycle regulators that help B cells progress through the cell cycle, and hence ensuring accurate variable-diversity-joining (VDJ) recombination and functional immune cell formation (By similarity). Together with ZFP36L2 is also necessary for thymocyte development and prevention of T-cell acute lymphoblastic leukemia (T-ALL) transformation by promoting ARE-mediated mRNA decay of the oncogenic transcription factor NOTCH1 mRNA (By similarity). Participates in the delivery of target ARE-mRNAs to processing bodies (PBs). In addition to its cytosolic mRNA-decay function, plays a role in the regulation of nuclear mRNA 3'-end processing; modulates mRNA 3'-end maturation efficiency of the DLL4 mRNA through binding with an ARE embedded in a weak noncanonical polyadenylation (poly(A)) signal in endothelial cells. Also involved in the regulation of stress granule (SG) and P-body (PB) formation and fusion. Plays a role in vasculogenesis and endocardial development (By similarity). Plays a role in the regulation of keratinocyte proliferation, differentiation and apoptosis. Plays a role in myoblast cell differentiation (By similarity).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MTTTLVSATIFDLSEVLCKGNKMLNYSAPSAGGCLLDRKAVGTPAGGGFPRRHSVTLPSS
KFHQNQLLSSLKGEPAPALSSRDSRFRDRSFSEGGERLLPTQKQPGGGQVNSSRYKTELC RPFEENGACKYGDKCQFAHGIHELRSLTRHPKYKTELCRTFHTIGFCPYGPRCHFIHNAE ERRALAGARDLSADRPRLQHSFSFAGFPSAAATAAATGLLDSPTSITPPPILSADDLLGS PTLPDGTNNPFAFSSQELASLFAPSMGLPGGGSPTTFLFRPMSESPHMFDSPPSPQDSLS DQEGYLSSSSSSHSGSDSPTLDNSRRLPIFSRLSISDD Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
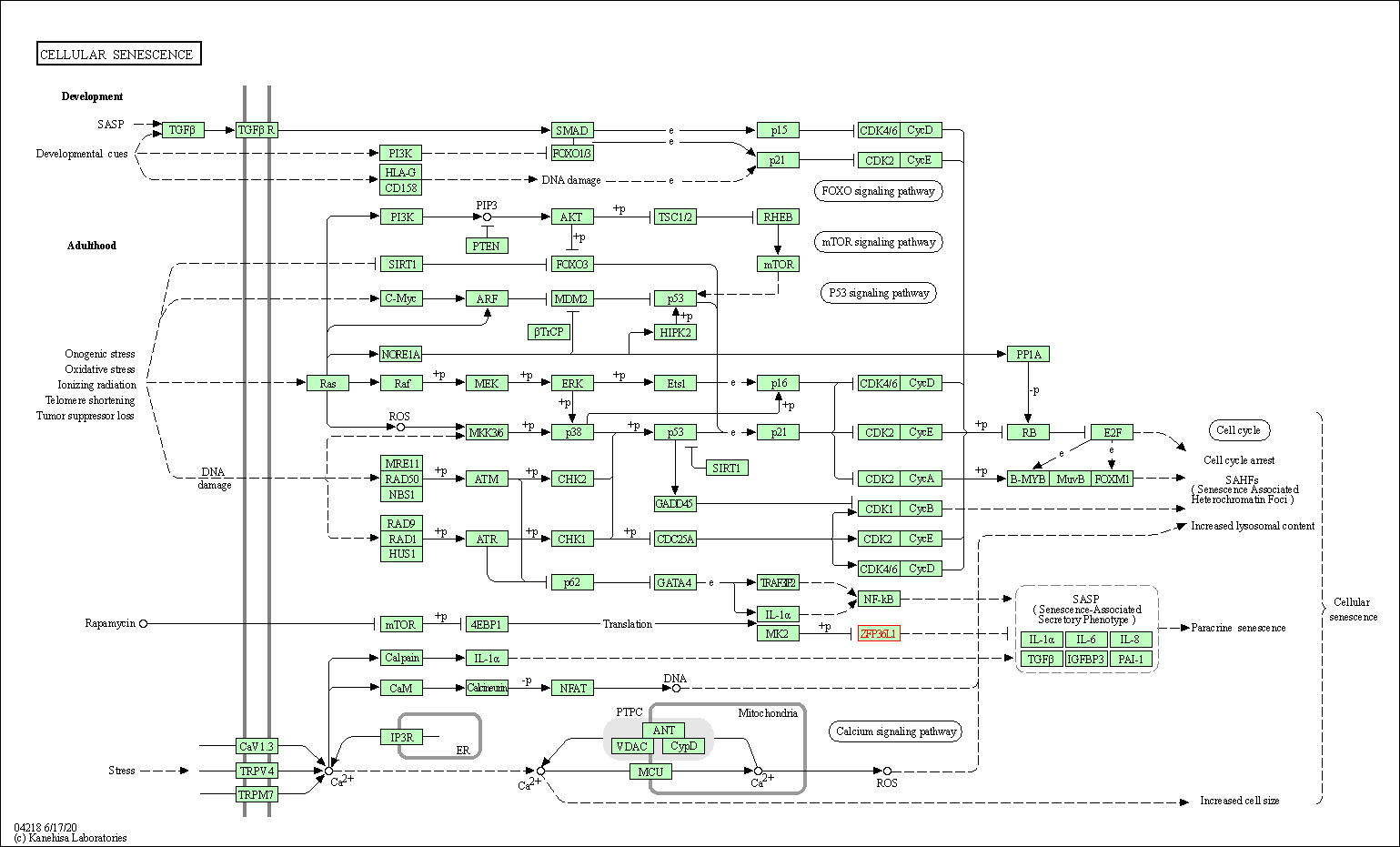
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cellular senescence | hsa04218 | Affiliated Target |

|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Degree | 1 | Degree centrality | 1.07E-04 | Betweenness centrality | 0.00E+00 |
|---|---|---|---|---|---|
| Closeness centrality | 1.76E-01 | Radiality | 1.29E+01 | Clustering coefficient | 0.00E+00 |
| Neighborhood connectivity | 8.00E+00 | Topological coefficient | 1.00E+00 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | ZFP36L1 negatively regulates erythroid differentiation of CD34+ hematopoietic stem cells by interfering with the Stat5b pathway. Mol Biol Cell. 2010 Oct 1;21(19):3340-51. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

