Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T38509
|
|||||
| Target Name |
Coagulation factor XII (F12)
|
|||||
| Synonyms |
Hageman factor; HAF
Click to Show/Hide
|
|||||
| Gene Name |
F12
|
|||||
| Target Type |
Clinical trial target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | Innate/adaptive immunodeficiency [ICD-11: 4A00] | |||||
| BioChemical Class |
Peptidase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 3.4.21.38
|
|||||
| Sequence |
MRALLLLGFLLVSLESTLSIPPWEAPKEHKYKAEEHTVVLTVTGEPCHFPFQYHRQLYHK
CTHKGRPGPQPWCATTPNFDQDQRWGYCLEPKKVKDHCSKHSPCQKGGTCVNMPSGPHCL CPQHLTGNHCQKEKCFEPQLLRFFHKNEIWYRTEQAAVARCQCKGPDAHCQRLASQACRT NPCLHGGRCLEVEGHRLCHCPVGYTGAFCDVDTKASCYDGRGLSYRGLARTTLSGAPCQP WASEATYRNVTAEQARNWGLGGHAFCRNPDNDIRPWCFVLNRDRLSWEYCDLAQCQTPTQ AAPPTPVSPRLHVPLMPAQPAPPKPQPTTRTPPQSQTPGALPAKREQPPSLTRNGPLSCG QRLRKSLSSMTRVVGGLVALRGAHPYIAALYWGHSFCAGSLIAPCWVLTAAHCLQDRPAP EDLTVVLGQERRNHSCEPCQTLAVRSYRLHEAFSPVSYQHDLALLRLQEDADGSCALLSP YVQPVCLPSGAARPSETTLCQVAGWGHQFEGAEEYASFLQEAQVPFLSLERCSAPDVHGS SILPGMLCAGFLEGGTDACQGDSGGPLVCEDQAAERRLTLQGIISWGSGCGDRNKPGVYT DVAYYLAWIREHTVS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| HIT2.0 ID | T21O8C | |||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Clinical Trial Drug(s) | [+] 2 Clinical Trial Drugs | + | ||||
| 1 | CSL312 | Drug Info | Phase 3 | Hereditary angioedema | [2] | |
| 2 | ION547 | Drug Info | Phase 1 | Thrombosis | [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 1 Inhibitor drugs | + | ||||
| 1 | CSL312 | Drug Info | [1] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Benzamidine | Ligand Info | |||||
| Structure Description | Structures of the two-chain human plasma Factor XIIa co-crystallized with potent inhibitors | PDB:6B74 | ||||
| Method | X-ray diffraction | Resolution | 2.32 Å | Mutation | No | [4] |
| PDB Sequence |
VVGGLVALRG
25 AHPYIAALYW37 GHSFCAGSLI47 APCWVLTAAH57 CLQDRPAPED63 LTVVLGQERR 73 NHSCEPCQTL83 AVRSYRLHEA93 FSPVSYQHDL103 ALLRLQEDAD109D GSCALLSPYV 118 QPVCLPSGAA128 RPSETTLCQV138 AGWGHQFEGA148 EEYASFLQEA158 QVPFLSLERC 168 SAPDVHGSSI176 LPGMLCAGFL185 EGGTDACQGD194 SGGPLVCEDQ204 AAERRLTLQG 211 IISWGSGCGD221A RNKPGVYTDV231 AYYLAWIREH241 TVS
|
|||||
|
|
||||||
| Ligand Name: US10875851, Example 114 | Ligand Info | |||||
| Structure Description | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h) | PDB:6X0T | ||||
| Method | X-ray diffraction | Resolution | 1.39 Å | Mutation | No | [5] |
| PDB Sequence |
LSCGQRLRVV
374 GGLVALRGAH384 PYIAALYWGH394 SFCAGSLIAP404 CWVLTAAHCL414 QDRPAPEDLT 424 VVLGQERRNH434 SCEPCQTLAV444 RSYRLHEAFS454 PVSYQHDLAL464 LRLQEDADGS 474 CALLSPYVQP484 VCLPSLCQVA503 GWGHQFEGAE513 EYASFLQEAQ523 VPFLSLERCS 533 APDVHGSSIL543 PGMLCAGFLE553 GGTDACQGDS563 GGPLVCEDQA573 AERRLTLQGI 583 ISWGSGCGDR593 NKPGVYTDVA603 YYLAWIREHT613 V
|
|||||
|
|
TRP392
3.887
SER395
4.328
PHE396
3.404
CYS397
3.477
HIS412
3.402
CYS413
3.290
PRO455
4.143
VAL456
3.695
SER457
4.651
TYR458
3.539
GLU510
3.324
TYR515
4.597
ASP557
4.914
ALA558
3.837
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Complement and coagulation cascades | hsa04610 | Affiliated Target |
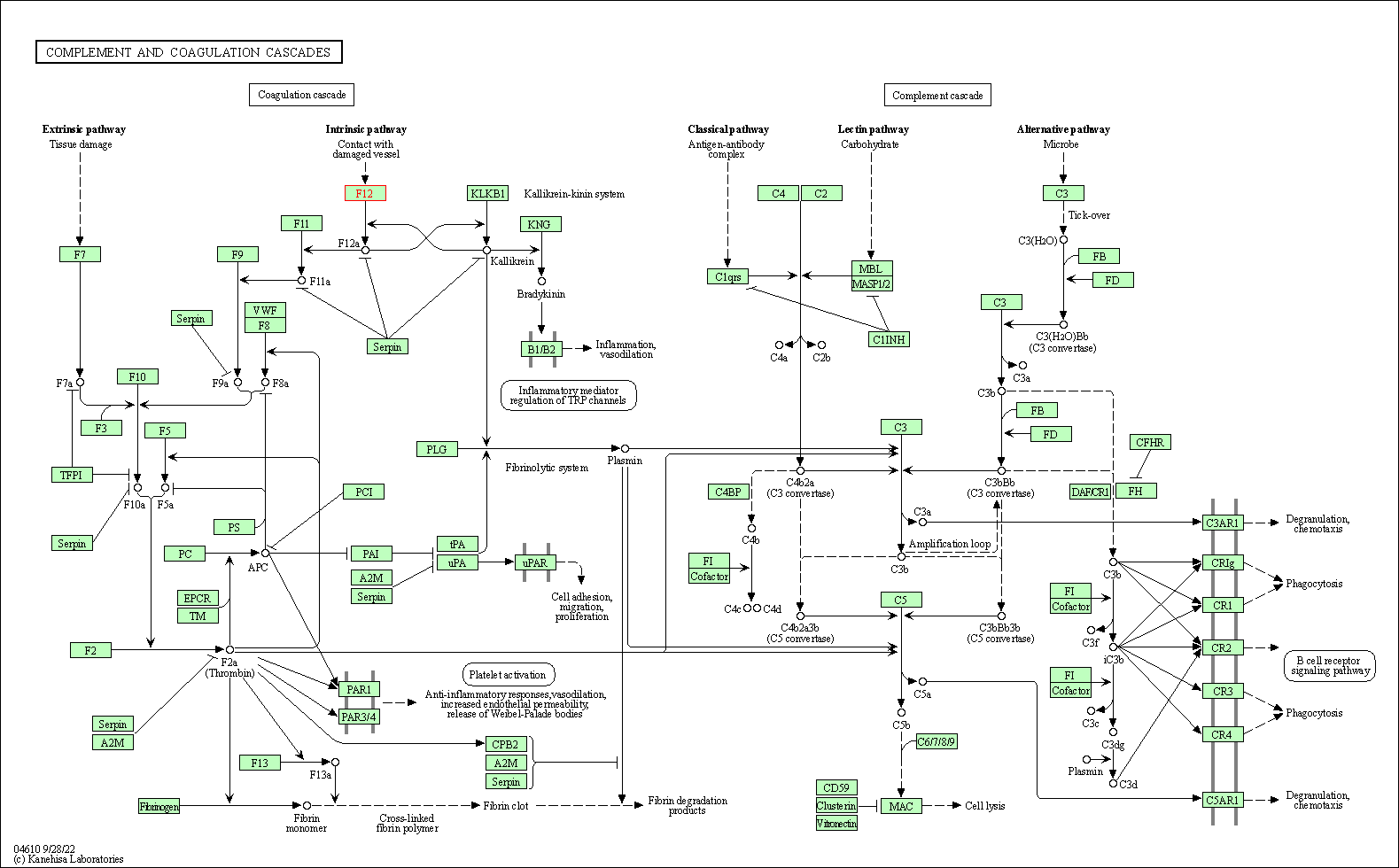
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 7 | Degree centrality | 7.52E-04 | Betweenness centrality | 1.61E-04 |
|---|---|---|---|---|---|
| Closeness centrality | 1.88E-01 | Radiality | 1.32E+01 | Clustering coefficient | 2.38E-01 |
| Neighborhood connectivity | 9.43E+00 | Topological coefficient | 1.98E-01 | Eccentricity | 13 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Clinical pipeline report, company report or official report of CSL Behring. | |||||
| REF 2 | ClinicalTrials.gov (NCT04656418) CSL312 (Garadacimab) in the Prevention of Hereditary Angioedema Attacks. U.S. National Institutes of Health. | |||||
| REF 3 | ClinicalTrials.gov (NCT04934891) A Double-Blind, Placebo-Controlled, Dose-Escalation Phase 1 Study to Assess the Safety, Tolerability, Pharmacokinetics and Pharmacodynamics of Single and Multiple Doses of ION547 Administered Subcutaneously to Healthy Subjects. U.S.National Institutes of Health. | |||||
| REF 4 | Structures of human plasma beta-factor XIIa cocrystallized with potent inhibitors. Blood Adv. 2018 Mar 13;2(5):549-558. | |||||
| REF 5 | Structure of human plasma factor XIIa in complex with (2S)-1-(N,3-dicyclohexyl-D-alanyl)-4-[(4R,5S)-4-methyl-5-phenyl-4,5-dihydro-1,3-oxazol-2-yl]-N-[(thiophen-2-yl)methyl]piperazine-2-carboxamide (compound 8h) | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

