Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T36292
(Former ID: TTDI03311)
|
|||||
| Target Name |
Calcium-activated potassium channel KCa5.1 (KCNU1)
|
|||||
| Synonyms |
Slowpoke homolog 3; SLO3; Potassium channel subfamily U member 1; KCa5; KCNMC1; KCNMA3; Calcium-activated potassium channel, subfamily M subunit alpha-3; Calcium-activated potassium channel subunit alpha-3
Click to Show/Hide
|
|||||
| Gene Name |
KCNU1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Testis-specific potassium channel activated by both intracellular pH and membrane voltage that mediates export of K(+). May represent the primary spermatozoan K(+) current. In contrast to KCNMA1/SLO1, it is not activated by Ca(2+) or Mg(2+). Critical for fertility. May play an important role in sperm osmoregulation required for the acquisition of normal morphology and motility when faced with osmotic challenges, such as those experienced after mixing with seminal fluid and entry into the vagina.
Click to Show/Hide
|
|||||
| BioChemical Class |
Voltage-gated ion channel
|
|||||
| UniProt ID | ||||||
| Sequence |
MFQTKLRNETWEDLPKMSCTTEIQAAFILSSFVTFFSGLIILLIFRLIWRSVKKWQIIKG
TGIILELFTSGTIARSHVRSLHFQGQFRDHIEMLLSAQTFVGQVLVILVFVLSIGSLIIY FINSADPVGSCSSYEDKTIPIDLVFNAFFSFYFGLRFMAADDKIKFWLEMNSIVDIFTIP PTFISYYLKSNWLGLRFLRALRLLELPQILQILRAIKTSNSVKFSKLLSIILSTWFTAAG FIHLVENSGDPWLKGRNSQNISYFESIYLVMATTSTVGFGDVVAKTSLGRTFIMFFTLGS LILFANYIPEMVELFANKRKYTSSYEALKGKKFIVVCGNITVDSVTAFLRNFLRDKSGEI NTEIVFLGETPPSLELETIFKCYLAYTTFISGSAMKWEDLRRVAVESAEACLIIANPLCS DSHAEDISNIMRVLSIKNYDSTTRIIIQILQSHNKVYLPKIPSWNWDTGDNIICFAELKL GFIAQGCLVPGLCTFLTSLFVEQNKKVMPKQTWKKHFLNSMKNKILTQRLSDDFAGMSFP EVARLCFLKMHLLLIAIEYKSLFTDGFCGLILNPPPQVRIRKNTLGFFIAETPKDVRRAL FYCSVCHDDVFIPELITNCGCKSRSRQHITVPSVKRMKKCLKGISSRISGQDSPPRVSAS TSSISNFTTRTLQHDVEQDSDQLDSSGMFHWCKPTSLDKVTLKRTGKSKYKFRNHIVACV FGDAHSAPMGLRNFVMPLRASNYTRKELKDIVFIGSLDYLQREWRFLWNFPQIYILPGCA LYSGDLHAANIEQCSMCAVLSPPPQPSSNQTLVDTEAIMATLTIGSLQIDSSSDPSPSVS EETPGYTNGHNEKSNCRKVPILTELKNPSNIHFIEQLGGLEGSLQETNLHLSTAFSTGTV FSGSFLDSLLATAFYNYHVLELLQMLVTGGVSSQLEQHLDKDKVYGVADSCTSLLSGRNR CKLGLLSLHETILSDVNPRNTFGQLFCGSLDLFGILCVGLYRIIDEEELNPENKRFVITR PANEFKLLPSDLVFCAIPFSTACYKRNEEFSLQKSYEIVNKASQTTETHSDTNCPPTIDS VTETLYSPVYSYQPRTNSLSFPKQIAWNQSRTNSIISSQIPLGDNAKENERKTSDEVYDE DPFAYSEPL Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
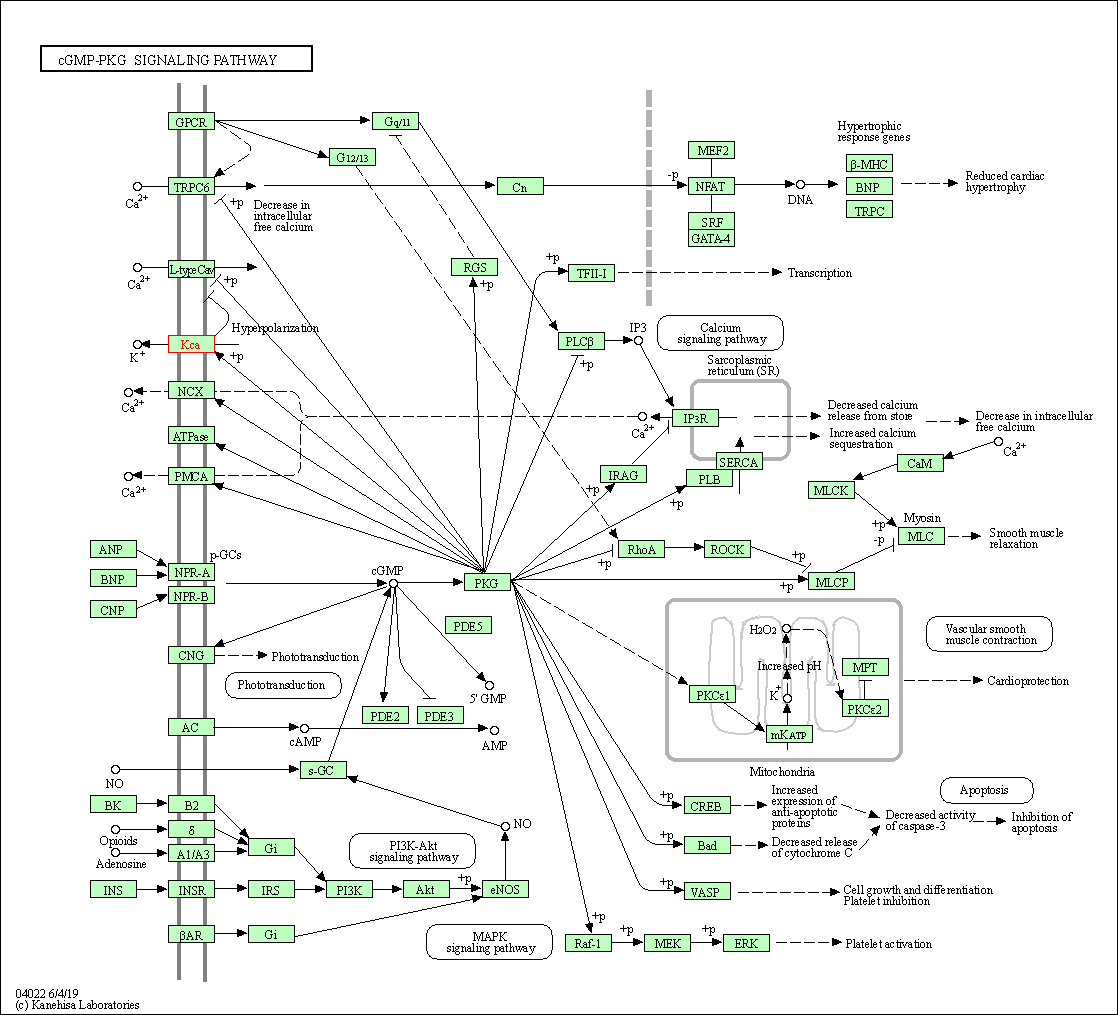
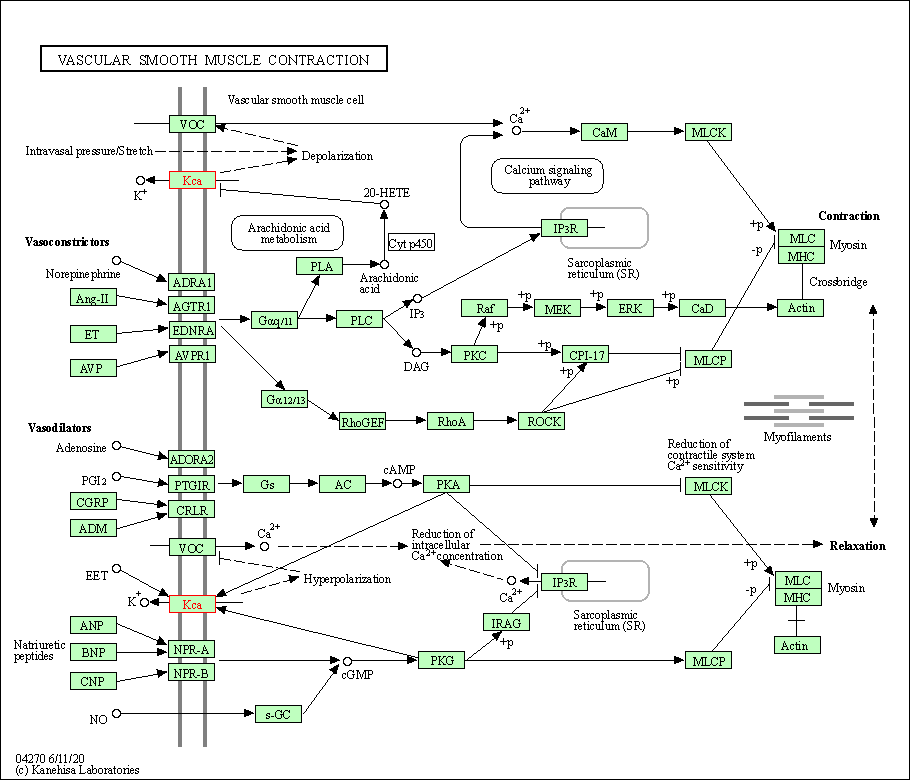
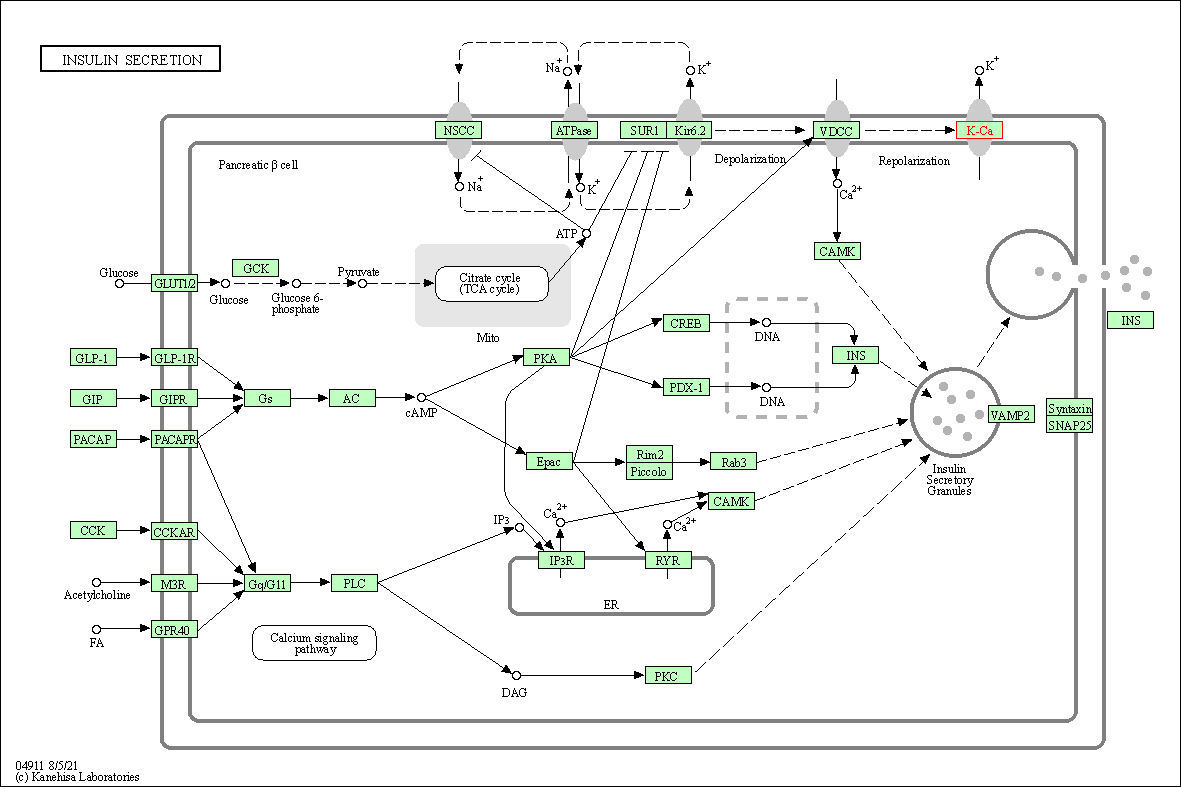
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| cGMP-PKG signaling pathway | hsa04022 | Affiliated Target |

|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |

|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Insulin secretion | hsa04911 | Affiliated Target |

|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 387). | |||||
| REF 2 | Block of mouse Slo1 and Slo3 K+ channels by CTX, IbTX, TEA, 4-AP and quinidine. Channels (Austin). 2010 Jan-Feb;4(1):22-41. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

