Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T23000
(Former ID: TTDI02079)
|
|||||
| Target Name |
Gelsolin (GSN)
|
|||||
| Synonyms |
GSN; Brevin; Actindepolymerizing factor; AGEL
Click to Show/Hide
|
|||||
| Gene Name |
GSN
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Calcium-regulated, actin-modulating protein that binds to the plus (or barbed) ends of actin monomers or filaments, preventing monomer exchange (end-blocking or capping). It can promote the assembly of monomers into filaments (nucleation) as well as sever filaments already formed. Plays a role in ciliogenesis. {ECO:0000269|PubMed:20393563}.
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MAPHRPAPALLCALSLALCALSLPVRAATASRGASQAGAPQGRVPEARPNSMVVEHPEFL
KAGKEPGLQIWRVEKFDLVPVPTNLYGDFFTGDAYVILKTVQLRNGNLQYDLHYWLGNEC SQDESGAAAIFTVQLDDYLNGRAVQHREVQGFESATFLGYFKSGLKYKKGGVASGFKHVV PNEVVVQRLFQVKGRRVVRATEVPVSWESFNNGDCFILDLGNNIHQWCGSNSNRYERLKA TQVSKGIRDNERSGRARVHVSEEGTEPEAMLQVLGPKPALPAGTEDTAKEDAANRKLAKL YKVSNGAGTMSVSLVADENPFAQGALKSEDCFILDHGKDGKIFVWKGKQANTEERKAALK TASDFITKMDYPKQTQVSVLPEGGETPLFKQFFKNWRDPDQTDGLGLSYLSSHIANVERV PFDAATLHTSTAMAAQHGMDDDGTGQKQIWRIEGSNKVPVDPATYGQFYGGDSYIILYNY RHGGRQGQIIYNWQGAQSTQDEVAASAILTAQLDEELGGTPVQSRVVQGKEPAHLMSLFG GKPMIIYKGGTSREGGQTAPASTRLFQVRANSAGATRAVEVLPKAGALNSNDAFVLKTPS AAYLWVGTGASEAEKTGAQELLRVLRAQPVQVAEGSEPDGFWEALGGKAAYRTSPRLKDK KMDAHPPRLFACSNKIGRFVIEEVPGELMQEDLATDDVMLLDTWDQVFVWVGKDSQEEEK TEALTSAKRYIETDPANRDRRTPITVVKQGFEPPSFVGWFLGWDDDYWSVDPLDRAMAEL AA Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T52GA6 | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: 2-(2-Methoxyethoxy)ethanol | Ligand Info | |||||
| Structure Description | HUMAN GELSOLIN FROM RESIDUES GLU28 TO ARG161 WITH CALCIUM | PDB:5ZZ0 | ||||
| Method | X-ray diffraction | Resolution | 2.63 Å | Mutation | No | [2] |
| PDB Sequence |
HPEFLKAGKE
38 PGLQIWRVEK48 FDLVPVPTNL58 YGDFFTGDAY68 VILKTVQLRN78 GNLQYDLHYW 88 LGNECSQDES98 GAAAIFTVQL108 DDYLNGRAVQ118 HREVQGFESA128 TFLGYFKSGL 138 KYKKGGVASG148 FK
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Fc gamma R-mediated phagocytosis | hsa04666 | Affiliated Target |
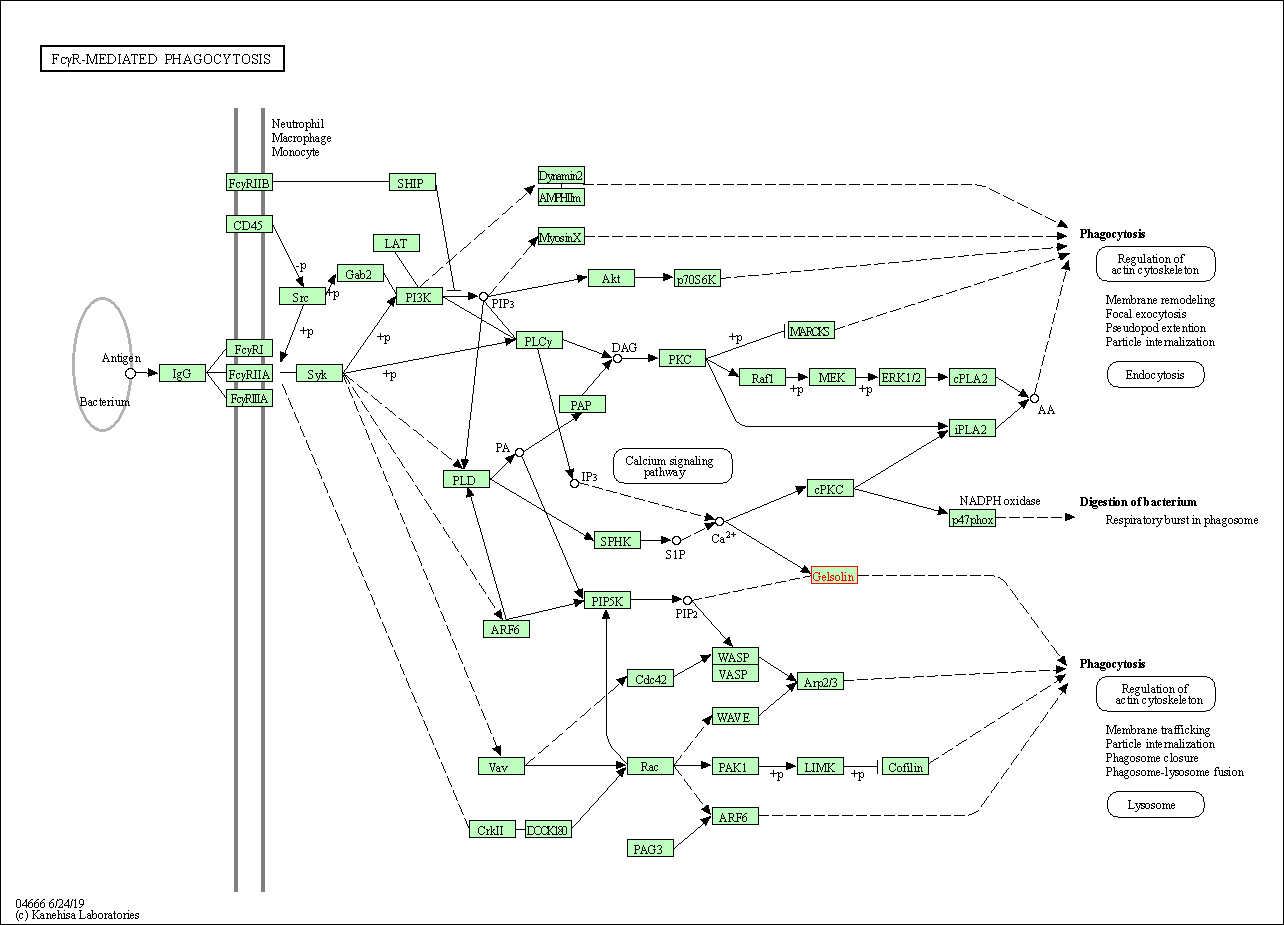
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Regulation of actin cytoskeleton | hsa04810 | Affiliated Target |
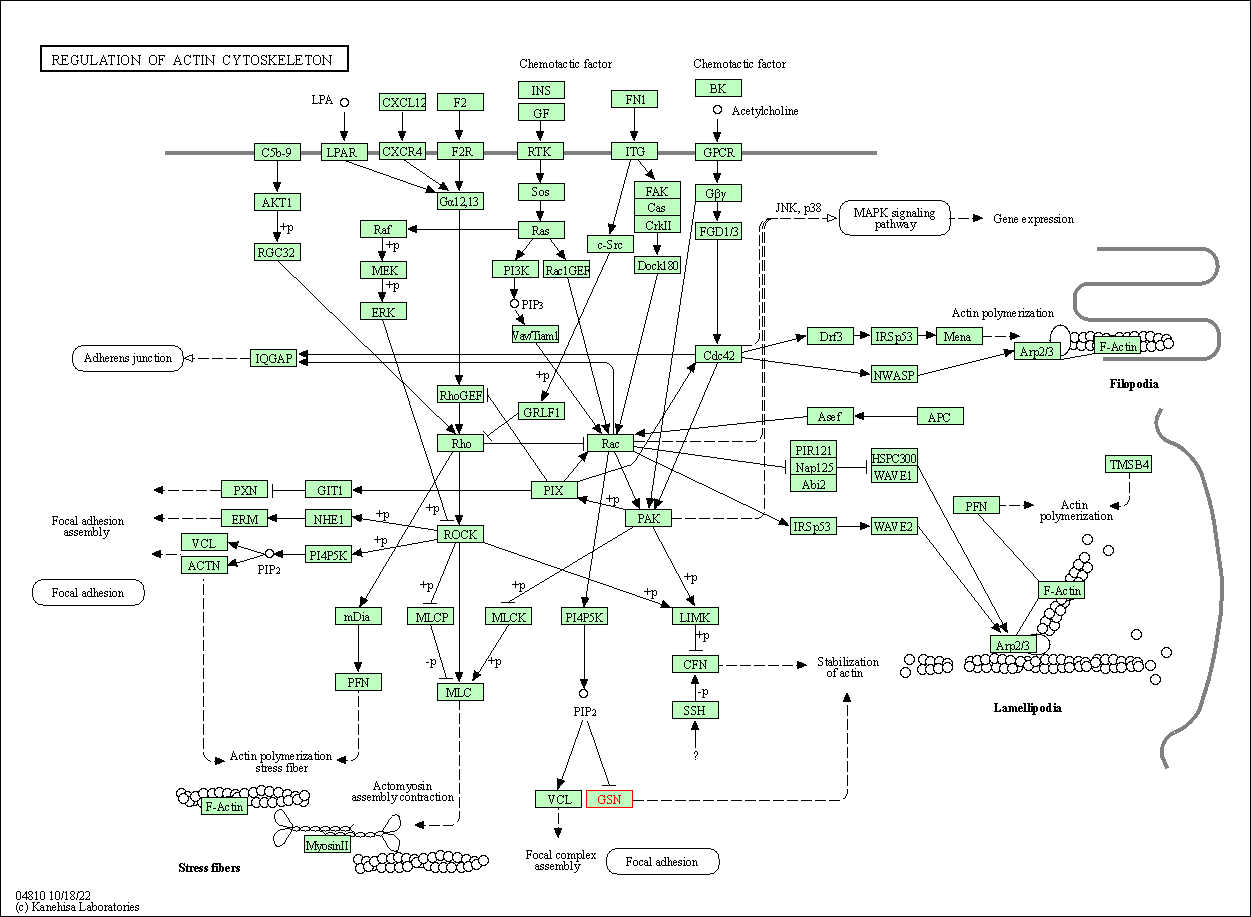
|
| Class: Cellular Processes => Cell motility | Pathway Hierarchy | ||
| Degree | 6 | Degree centrality | 6.45E-04 | Betweenness centrality | 3.41E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.29E-01 | Radiality | 1.40E+01 | Clustering coefficient | 2.67E-01 |
| Neighborhood connectivity | 5.65E+01 | Topological coefficient | 2.08E-01 | Eccentricity | 12 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 3 KEGG Pathways | + | ||||
| 1 | Fc gamma R-mediated phagocytosis | |||||
| 2 | Regulation of actin cytoskeleton | |||||
| 3 | Viral carcinogenesis | |||||
| Panther Pathway | [+] 1 Panther Pathways | + | ||||
| 1 | FAS signaling pathway | |||||
| PID Pathway | [+] 5 PID Pathways | + | ||||
| 1 | Coregulation of Androgen receptor activity | |||||
| 2 | Osteopontin-mediated events | |||||
| 3 | EGF receptor (ErbB1) signaling pathway | |||||
| 4 | Caspase Cascade in Apoptosis | |||||
| 5 | N-cadherin signaling events | |||||
| Reactome | [+] 2 Reactome Pathways | + | ||||
| 1 | Caspase-mediated cleavage of cytoskeletal proteins | |||||
| 2 | Amyloid formation | |||||
| WikiPathways | [+] 3 WikiPathways | + | ||||
| 1 | Senescence and Autophagy in Cancer | |||||
| 2 | Regulation of Actin Cytoskeleton | |||||
| 3 | Apoptotic execution phase | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Gelsolin as therapeutic target in Alzheimer's disease. Expert Opin Ther Targets. 2010 Jun;14(6):585-92. | |||||
| REF 2 | Bonsai Gelsolin Survives Heat Induced Denaturation by Forming beta-Amyloids which Leach Out Functional Monomer. Sci Rep. 2018 Aug 22;8(1):12602. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

