Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T19615
(Former ID: TTDR01234)
|
|||||
| Target Name |
Granulocyte colony-stimulating factor (CSF3)
|
|||||
| Synonyms |
Pluripoietin; Lenograstim; G-CSF; CSF3
Click to Show/Hide
|
|||||
| Gene Name |
CSF3
|
|||||
| Target Type |
Successful target
|
[1] | ||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Aplastic anaemia [ICD-11: 3A70] | |||||
| 2 | Neutropenia [ICD-11: 4B00] | |||||
| Function |
Granulocyte/macrophage colony-stimulating factors are cytokines that act in hematopoiesis by controlling the production, differentiation, and function of 2 related white cell populations of the blood, the granulocytes and the monocytes-macrophages. This CSF induces granulocytes.
Click to Show/Hide
|
|||||
| BioChemical Class |
Cytokine: interleukin
|
|||||
| UniProt ID | ||||||
| Sequence |
MAGPATQSPMKLMALQLLLWHSALWTVQEATPLGPASSLPQSFLLKCLEQVRKIQGDGAA
LQEKLVSECATYKLCHPEELVLLGHSLGIPWAPLSSCPSQALQLAGCLSQLHSGLFLYQG LLQALEGISPELGPTLDTLQLDVADFATTIWQQMEELGMAPALQPTQGAMPAFASAFQRR AGGVLVASHLQSFLEVSYRVLRHLAQP Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Approved Drug(s) | [+] 2 Approved Drugs | + | ||||
| 1 | Efbemalenograstim | Drug Info | Approved | Neutropenia | [2] | |
| 2 | Interleukin-3 | Drug Info | Approved | Aplastic anemia | [3] | |
| Clinical Trial Drug(s) | [+] 4 Clinical Trial Drugs | + | ||||
| 1 | Neugranin | Drug Info | Preregistration | Neutropenia | [4] | |
| 2 | DA-3031 | Drug Info | Phase 3 | Neutropenia | [5] | |
| 3 | LA-EP2006 | Drug Info | Phase 3 | Neutropenia | [6] | |
| 4 | MK-4214 | Drug Info | Phase 3 | Neutropenia | [7] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Modulator | [+] 6 Modulator drugs | + | ||||
| 1 | Efbemalenograstim | Drug Info | [2] | |||
| 2 | Interleukin-3 | Drug Info | [1], [3] | |||
| 3 | Neugranin | Drug Info | [8] | |||
| 4 | DA-3031 | Drug Info | [9], [10] | |||
| 5 | LA-EP2006 | Drug Info | [11] | |||
| 6 | MK-4214 | Drug Info | [12] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Cytokine-cytokine receptor interaction | hsa04060 | Affiliated Target |
|
| Class: Environmental Information Processing => Signaling molecules and interaction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Hematopoietic cell lineage | hsa04640 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
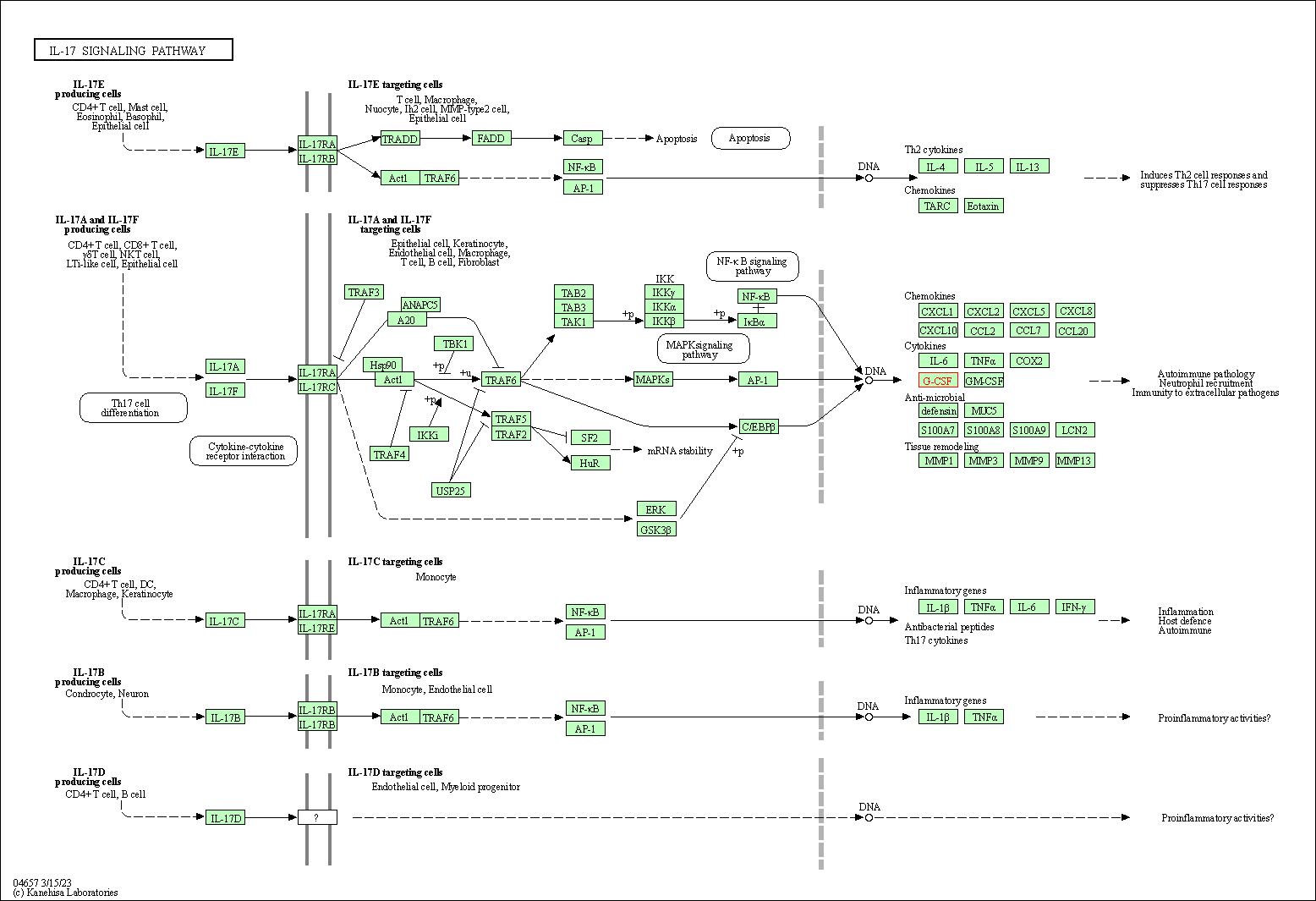
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Degree | 23 | Degree centrality | 2.47E-03 | Betweenness centrality | 5.79E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.16E-01 | Radiality | 1.38E+01 | Clustering coefficient | 5.22E-01 |
| Neighborhood connectivity | 2.57E+01 | Topological coefficient | 1.31E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Target Profiles in Patients | Top | |||||
|---|---|---|---|---|---|---|
| Target Expression Profile (TEP) | ||||||
| Target Affiliated Biological Pathways | Top | |||||
|---|---|---|---|---|---|---|
| KEGG Pathway | [+] 5 KEGG Pathways | + | ||||
| 1 | Cytokine-cytokine receptor interaction | |||||
| 2 | PI3K-Akt signaling pathway | |||||
| 3 | Jak-STAT signaling pathway | |||||
| 4 | Hematopoietic cell lineage | |||||
| 5 | Malaria | |||||
| NetPath Pathway | [+] 3 NetPath Pathways | + | ||||
| 1 | IL1 Signaling Pathway | |||||
| 2 | TNFalpha Signaling Pathway | |||||
| 3 | TCR Signaling Pathway | |||||
| WikiPathways | [+] 2 WikiPathways | + | ||||
| 1 | Cytokines and Inflammatory Response | |||||
| 2 | Hematopoietic Stem Cell Differentiation | |||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Interleukin 3: from colony-stimulating factor to pluripotent immunoregulatory cytokine. Int J Immunopharmacol. 1992 Apr;14(3):421-30. | |||||
| REF 2 | FDA Approved Drug Products from FDA Official Website. 2023. Application Number: 761134 | |||||
| REF 3 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 4 | Trusted, scientifically sound profiles of drug programs, clinical trials, safety reports, and company deals, written by scientists. Springer. 2015. Adis Insight (drug id 800017176) | |||||
| REF 5 | ClinicalTrials.gov (NCT01674855) Phase III Study of DA-3031(PEG-G-CSF) in Chemotherapy-induced Neutropenia. U.S. National Institutes of Health. | |||||
| REF 6 | ClinicalTrials.gov (NCT01516736) Phase III Study Comparing the Efficacy and Safety of LA-EP2006 and Peg-Filgrastim. U.S. National Institutes of Health. | |||||
| REF 7 | Clinical pipeline report, company report or official report of Merck. | |||||
| REF 8 | Development and Characterization of a Novel Fusion Protein of a Mutated Granulocyte Colony-Stimulating Factor and Human Serum Albumin in Pichia pastoris. PLoS One. 2014; 9(12): e115840. | |||||
| REF 9 | A randomized, multi-center, open-label, phase II study of once-per-cycle DA-3031, a biosimilar pegylated G-CSF, compared with daily filgrastim in patients receiving TAC chemotherapy for early-stage breast cancer.Invest New Drugs.2013 Oct;31(5):1300-6. | |||||
| REF 10 | Relationship between absolute neutrophil count profiles and pharmacokinetics of DA-3031, a pegylated granulocyte colony-stimulating factor (pegylated-G-CSF): a dose block-randomized, double-blind, dose-escalation study in healthy subjects. Clin Drug Investig. 2013 Nov;33(11):817-24. | |||||
| REF 11 | National Cancer Institute Drug Dictionary (drug id 724518). | |||||
| REF 12 | DOI: 10.1038/nbt0710-636a | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

