Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T18978
(Former ID: TTDI03601)
|
|||||
| Target Name |
Polycystic kidney disease 2-like 1 (TRPP2)
|
|||||
| Synonyms |
Polycystin-L1; Polycystin-L; Polycystin-2L1; Polycystin-2 homolog; Polycystic kidney disease 2-like 1 protein; PKDL; PKD2L
Click to Show/Hide
|
|||||
| Gene Name |
PKD2L1
|
|||||
| Target Type |
Literature-reported target
|
[1] | ||||
| Function |
Pore-forming subunit of a heterotetrameric, non-selective cation channel that is permeable to Ca(2+). Pore-forming subunit of a calcium-permeant ion channel formed by PKD1L2 and PKD1L1 in primary cilia, where it controls cilium calcium concentration, but does not affect cytoplasmic calcium concentration. The channel formed by PKD1L2 and PKD1L1 in primary cilia regulates sonic hedgehog/SHH signaling and GLI2 transcription. Pore-forming subunit of a channel formed by PKD1L2 and PKD1L3 that contributes to sour taste perception in gustatory cells. The heteromeric channel formed by PKD1L2 and PKD1L3 is activated by low pH, but opens only when the extracellular pH rises again. May play a role in the perception of carbonation taste (By similarity). May play a role in the sensory perception of water, via a mechanism that activates the channel in response to dilution of salivary bicarbonate and changes in salivary pH (By similarity).
Click to Show/Hide
|
|||||
| UniProt ID | ||||||
| Sequence |
MNAVGSPEGQELQKLGSGAWDNPAYSGPPSPHGTLRVCTISSTGPLQPQPKKPEDEPQET
AYRTQVSSCCLHICQGIRGLWGTTLTENTAENRELYIKTTLRELLVYIVFLVDICLLTYG MTSSSAYYYTKVMSELFLHTPSDTGVSFQAISSMADFWDFAQGPLLDSLYWTKWYNNQSL GHGSHSFIYYENMLLGVPRLRQLKVRNDSCVVHEDFREDILSCYDVYSPDKEEQLPFGPF NGTAWTYHSQDELGGFSHWGRLTSYSGGGYYLDLPGSRQGSAEALRALQEGLWLDRGTRV VFIDFSVYNANINLFCVLRLVVEFPATGGAIPSWQIRTVKLIRYVSNWDFFIVGCEVIFC VFIFYYVVEEILELHIHRLRYLSSIWNILDLVVILLSIVAVGFHIFRTLEVNRLMGKLLQ QPNTYADFEFLAFWQTQYNNMNAVNLFFAWIKIFKYISFNKTMTQLSSTLARCAKDILGF AVMFFIVFFAYAQLGYLLFGTQVENFSTFIKCIFTQFRIILGDFDYNAIDNANRILGPAY FVTYVFFVFFVLLNMFLAIINDTYSEVKEELAGQKDELQLSDLLKQGYNKTLLRLRLRKE RVSDVQKVLQGGEQEIQFEDFTNTLRELGHAEHEITELTATFTKFDRDGNRILDEKEQEK MRQDLEEERVALNTEIEKLGRSIVSSPQGKSGPEAARAGGWVSGEEFYMLTRRVLQLETV LEGVVSQIDAVGSKLKMLERKGWLAPSPGVKEQAIWKHPQPAPAVTPDPWGVQGGQESEV PYKREEEALEERRLSRGEIPTLQRS Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Human Similarity Proteins
Human Pathway Affiliation
|
|
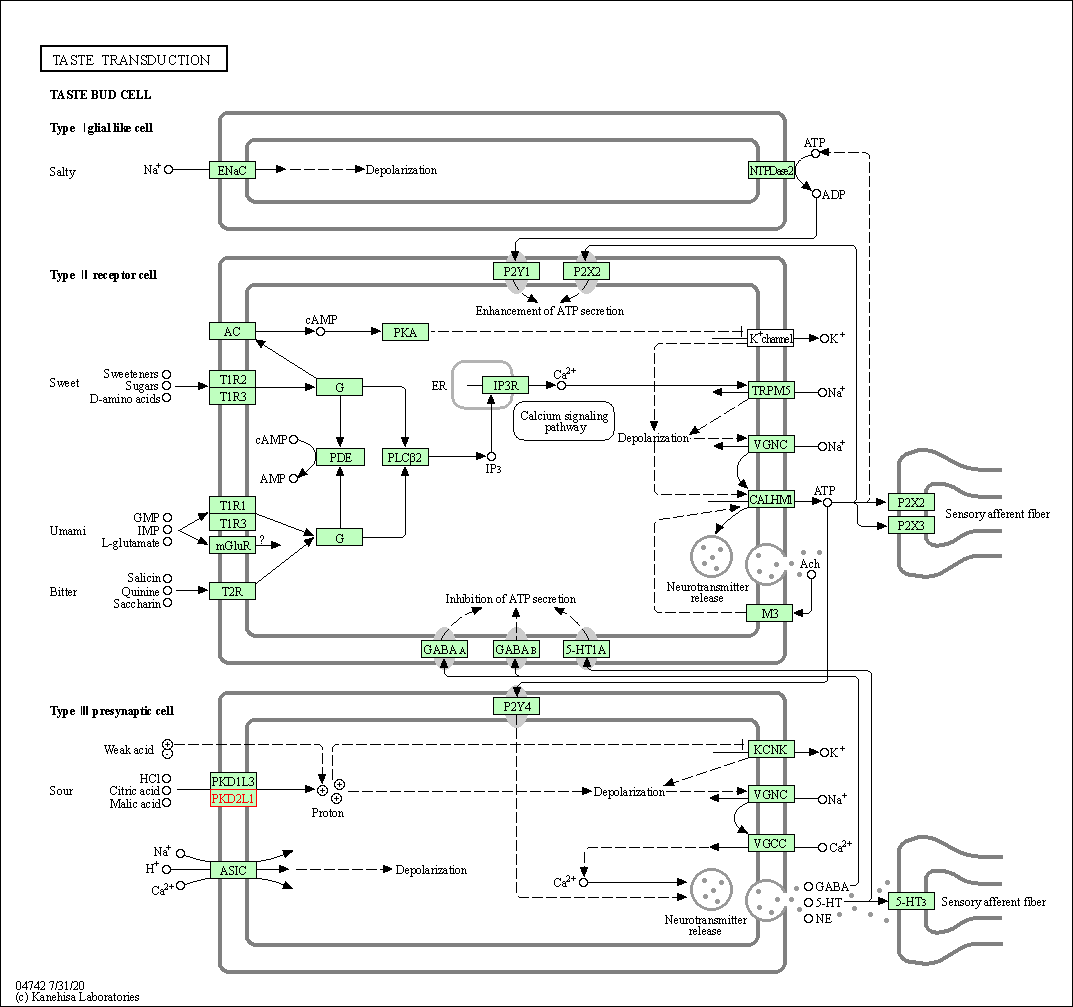
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| Taste transduction | hsa04742 | Affiliated Target |

|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 505). | |||||
| REF 2 | Transient receptor potential family members PKD1L3 and PKD2L1 form a candidate sour taste receptor. Proc Natl Acad Sci U S A. 2006 Aug 15;103(33):12569-74. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

