Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T00663
(Former ID: TTDR00928)
|
|||||
| Target Name |
Stress-activated protein kinase JNK3 (JNK3)
|
|||||
| Synonyms |
Stress-activated protein kinase 1b; SAPK1b; PRKM10; Mitogen-activated protein kinase 10; MAPK 10; MAP kinase p49 3F12; MAP kinase 10; JNK3A; C-Jun N-terminal kinase 3
Click to Show/Hide
|
|||||
| Gene Name |
MAPK10
|
|||||
| Target Type |
Patented-recorded target
|
[1] | ||||
| Disease | [+] 1 Target-related Diseases | + | ||||
| 1 | CNS birth injury [ICD-11: KA40] | |||||
| Function |
Extracellular stimuli such as proinflammatory cytokines or physical stress stimulate the stress-activated protein kinase/c-Jun N-terminal kinase (SAP/JNK) signaling pathway. In this cascade, two dual specificity kinases MAP2K4/MKK4 and MAP2K7/MKK7 phosphorylate and activate MAPK10/JNK3. In turn, MAPK10/JNK3 phosphorylates a number of transcription factors, primarily components of AP-1 such as JUN and ATF2 and thus regulates AP-1 transcriptional activity. Plays regulatory roles in the signaling pathways during neuronal apoptosis. Phosphorylates the neuronal microtubule regulator STMN2. Acts in the regulation of the amyloid-beta precursor protein/APP signaling during neuronal differentiation by phosphorylating APP. Participates also in neurite growth in spiral ganglion neurons. Phosphorylates the CLOCK-ARNTL/BMAL1 heterodimer and plays a role in the photic regulation of the circadian clock. Serine/threonine-protein kinase involved in various processes such as neuronal proliferation, differentiation, migration and programmed cell death.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.24
|
|||||
| Sequence |
MSLHFLYYCSEPTLDVKIAFCQGFDKQVDVSYIAKHYNMSKSKVDNQFYSVEVGDSTFTV
LKRYQNLKPIGSGAQGIVCAAYDAVLDRNVAIKKLSRPFQNQTHAKRAYRELVLMKCVNH KNIISLLNVFTPQKTLEEFQDVYLVMELMDANLCQVIQMELDHERMSYLLYQMLCGIKHL HSAGIIHRDLKPSNIVVKSDCTLKILDFGLARTAGTSFMMTPYVVTRYYRAPEVILGMGY KENVDIWSVGCIMGEMVRHKILFPGRDYIDQWNKVIEQLGTPCPEFMKKLQPTVRNYVEN RPKYAGLTFPKLFPDSLFPADSEHNKLKASQARDLLSKMLVIDPAKRISVDDALQHPYIN VWYDPAEVEAPPPQIYDKQLDEREHTIEEWKELIYKEVMNSEEKTKNGVVKGQPSPSGAA VNSSESLPPSSSVNDISSMSTDQTLASDTDSSLEASAGPLGCCR Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| HIT2.0 ID | T04WTG | |||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine monophosphate | Ligand Info | |||||
| Structure Description | The crystal structure of AMP-bound JNK3 | PDB:4KKE | ||||
| Method | X-ray diffraction | Resolution | 2.20 Å | Mutation | No | [10] |
| PDB Sequence |
DNQFYSVEVG
54 DSTFTVLKRY64 QNLKPIGSGA74 QGIVCAAYDA84 VLDRNVAIKK94 LSRPFQNQTH 104 AKRAYRELVL114 MKCVNHKNII124 SLLNVFTPQK134 TLEEFQDVYL144 VMELMDANLC 154 QVIQMELDHE164 RMSYLLYQML174 CGIKHLHSAG184 IIHRDLKPSN194 IVVKSDCTLK 204 ILDFGLARTA214 SFMMTPYVVT226 RYYRAPEVIL236 GMGYKENVDI246 WSVGCIMGEM 256 VRHKILFPGR266 DYIDQWNKVI276 EQLGTPCPEF286 MKKLQPTVRN296 YVENRPKYAG 306 LTFPKLFPDS316 LFPADSEHNK326 LKASQARDLL336 SKMLVIDPAK346 RISVDDALQH 356 PYINVWYDPA366 EVEAPPPQIY376 DKQLDEREHT386 IEEWKELIYK396 EVMN |
|||||
|
|
ILE70
3.497
GLY71
3.619
SER72
3.369
GLY73
3.426
ALA74
4.231
GLY76
4.844
VAL78
3.602
ALA91
3.548
LYS93
3.029
ILE124
4.077
MET146
3.834
|
|||||
| Ligand Name: CC-930 | Ligand Info | |||||
| Structure Description | Crystal Structure of JNK3 complexed with CC-930, an orally active anti-fibrotic JNK inhibitor | PDB:3TTI | ||||
| Method | X-ray diffraction | Resolution | 2.20 Å | Mutation | No | [11] |
| PDB Sequence |
NQFYSVEVGD
55 STFTVLKRYQ65 NLKPIGSGAQ75 GIVCAAYDAV85 LDRNVAIKKL95 SRPFQNQTHA 105 KRAYRELVLM115 KCVNHKNIIS125 LLNVFTPQKT135 LEEFQDVYLV145 MELMDANLCQ 155 VIQMELDHER165 MSYLLYQMLC175 GIKHLHSAGI185 IHRDLKPSNI195 VVKSDCTLKI 205 LDFGLASFMM220 TPYVVTRYYR230 APEVILGMGY240 KENVDIWSVG250 CIMGEMVRHK 260 ILFPGRDYID270 QWNKVIEQLG280 TPCPEFMKKL290 QPTVRNYVEN300 RPKYAGLTFP 310 KLFPDSLFPA320 DSEHNKLKAS330 QARDLLSKML340 VIDPAKRISV350 DDALQHPYIN 360 VWYDPAEVEA370 PPPREHTIEE389 WKELIYKEVM399 N
|
|||||
|
|
ILE70
3.652
GLY71
3.748
VAL78
3.482
ALA91
3.308
ILE92
4.065
LYS93
3.285
MET115
3.630
ILE124
3.293
LEU126
3.547
LEU144
3.373
VAL145
4.887
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
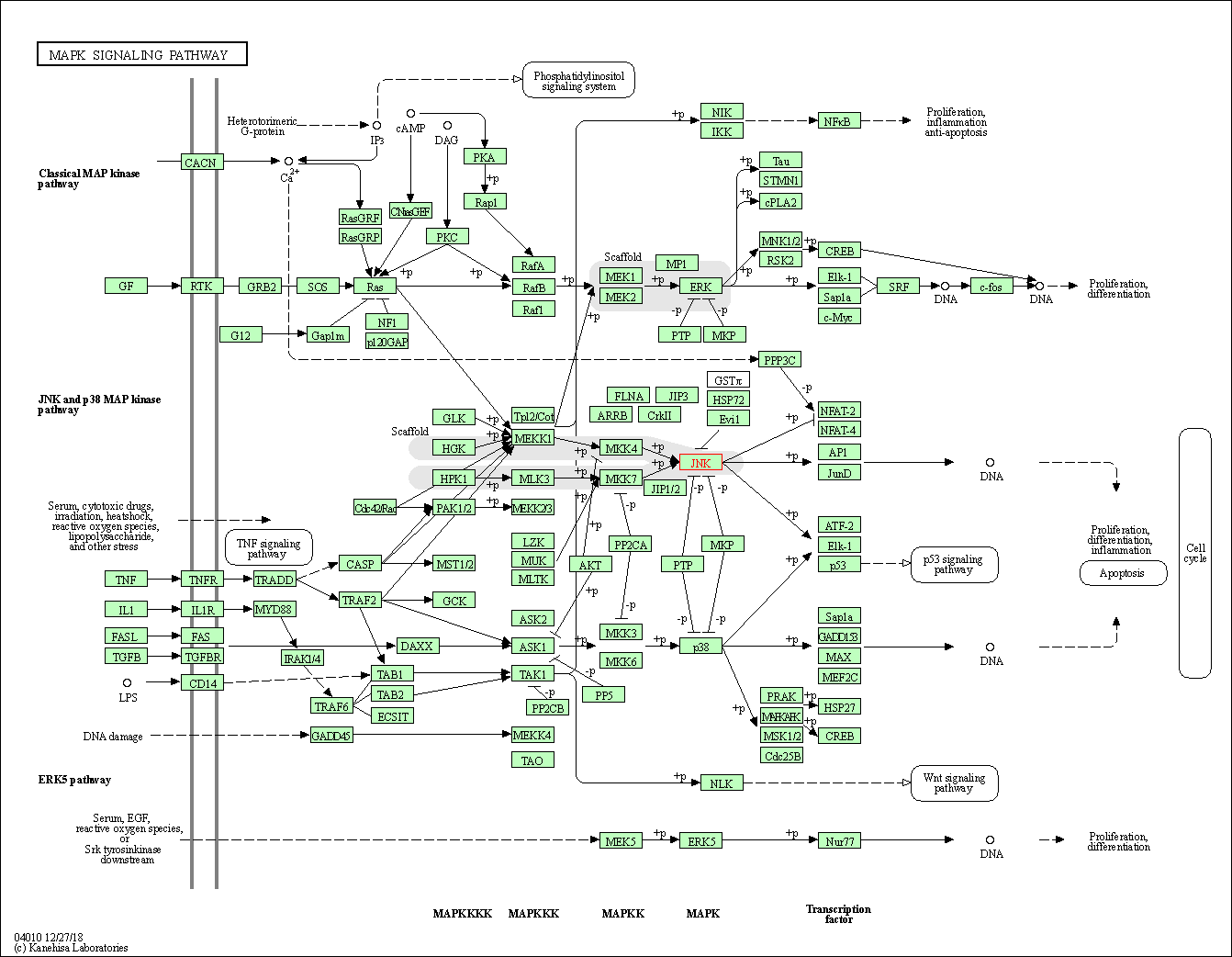
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| ErbB signaling pathway | hsa04012 | Affiliated Target |
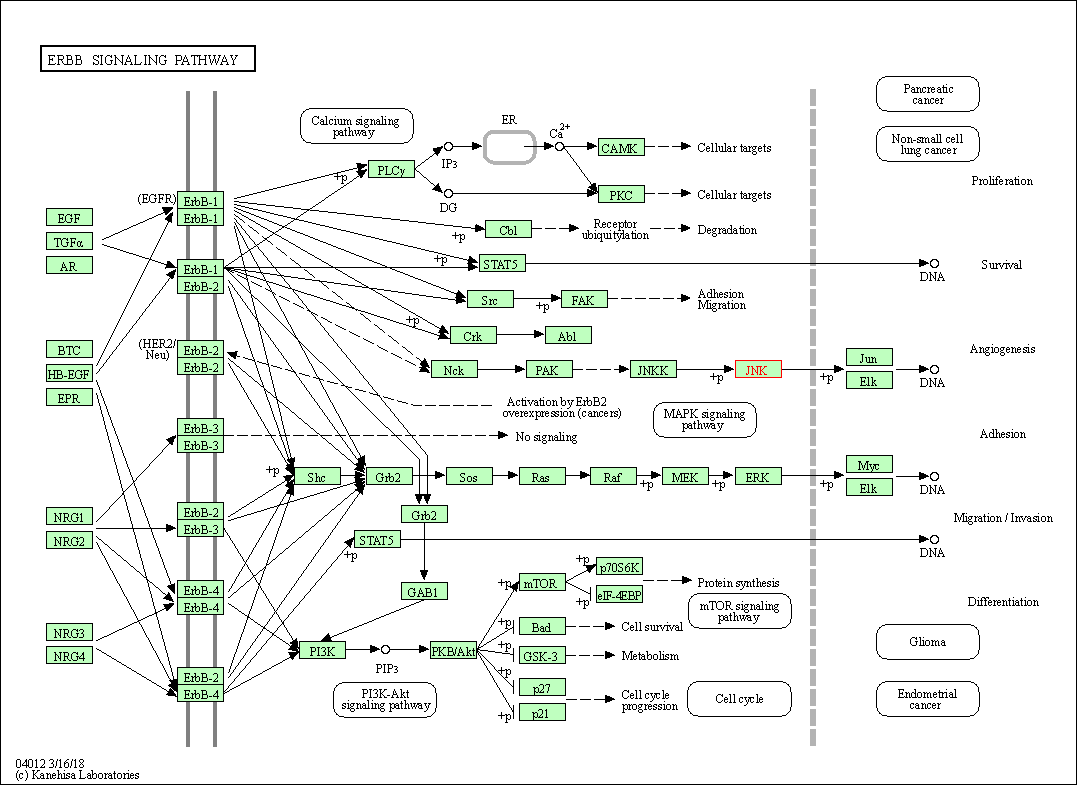
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |
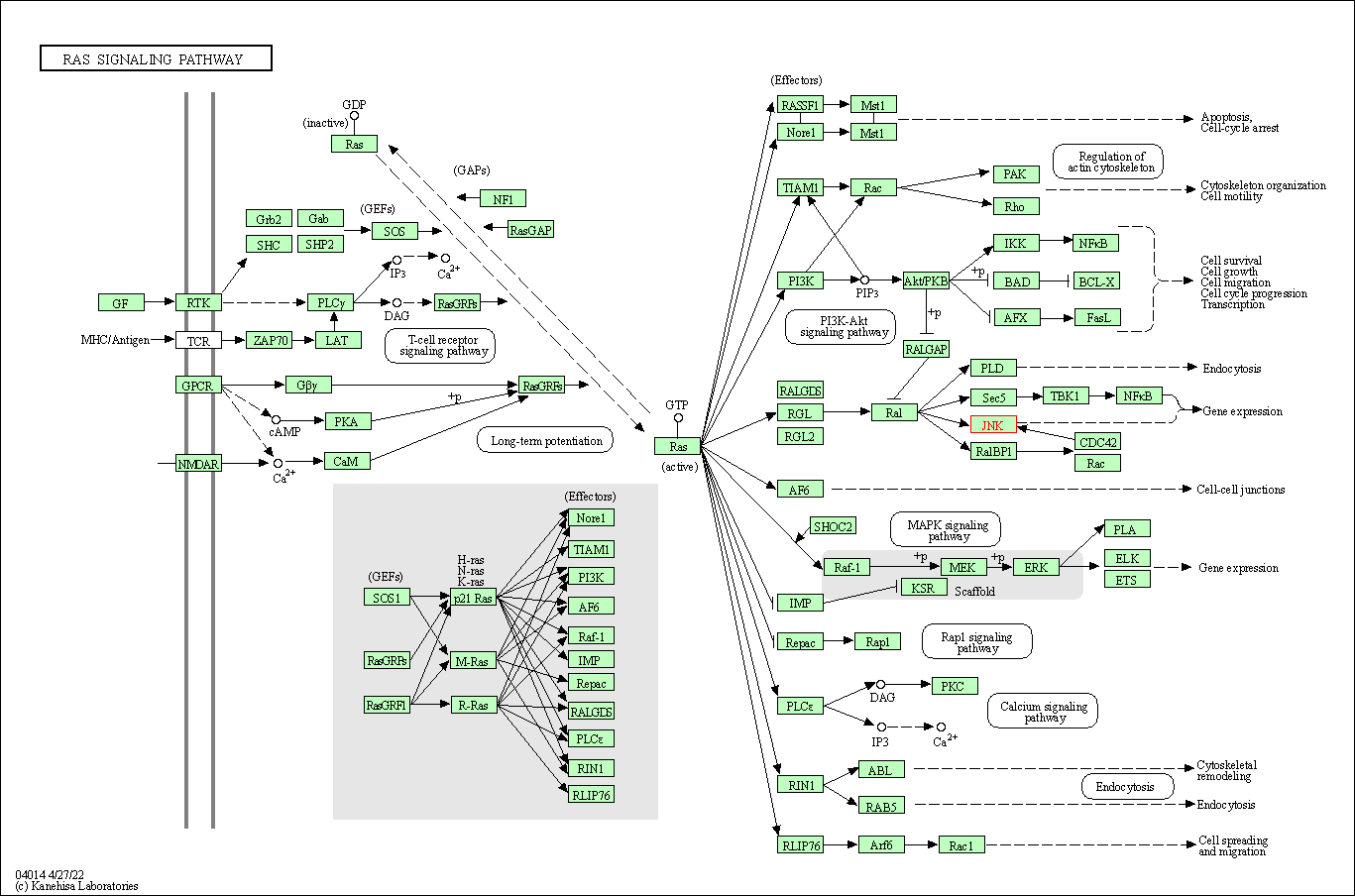
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
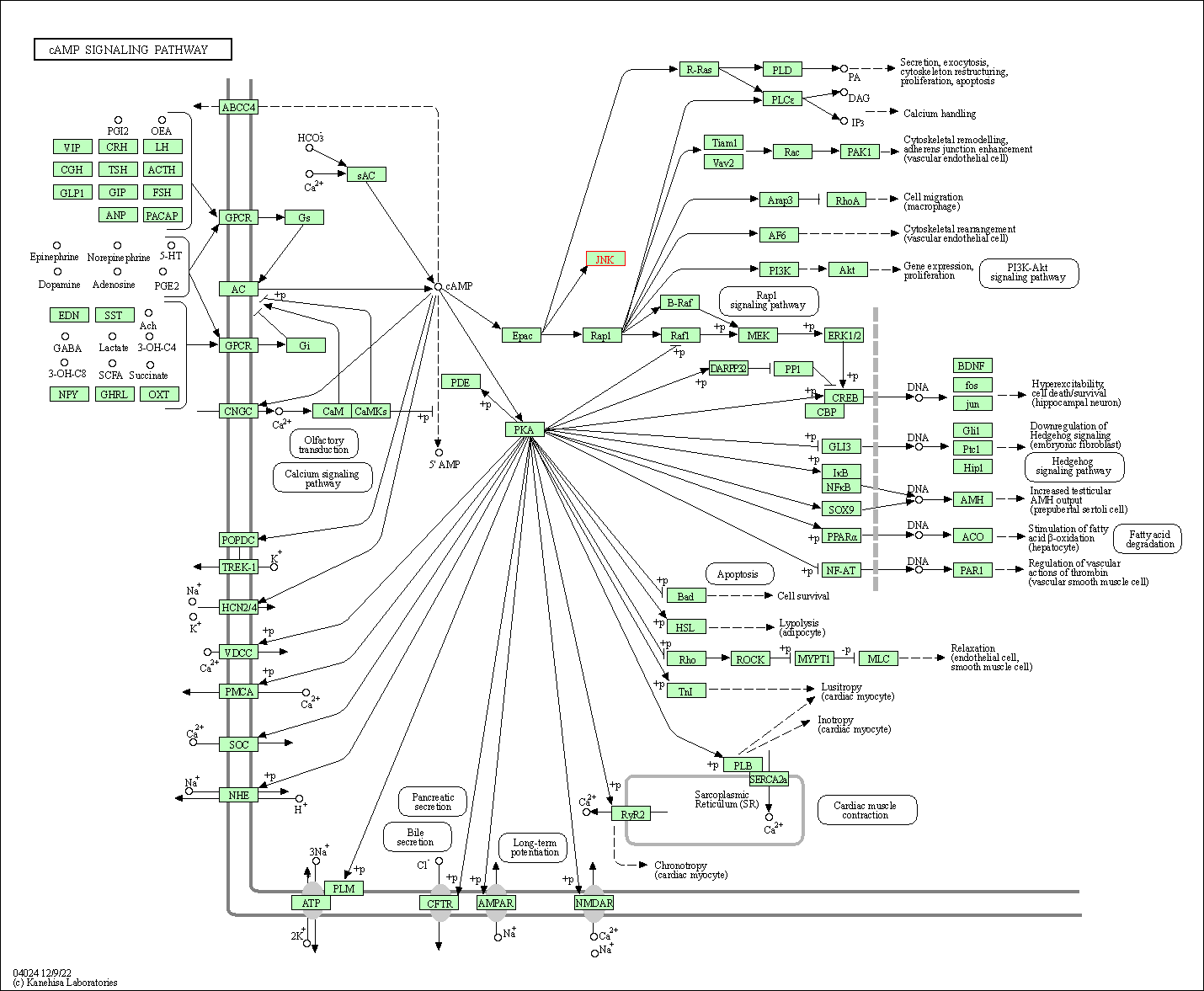
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| FoxO signaling pathway | hsa04068 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Sphingolipid signaling pathway | hsa04071 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Mitophagy - animal | hsa04137 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Protein processing in endoplasmic reticulum | hsa04141 | Affiliated Target |
|
| Class: Genetic Information Processing => Folding, sorting and degradation | Pathway Hierarchy | ||
| Apoptosis | hsa04210 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Apoptosis - multiple species | hsa04215 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Necroptosis | hsa04217 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Wnt signaling pathway | hsa04310 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Osteoclast differentiation | hsa04380 | Affiliated Target |
|
| Class: Organismal Systems => Development and regeneration | Pathway Hierarchy | ||
| Focal adhesion | hsa04510 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Toll-like receptor signaling pathway | hsa04620 | Affiliated Target |
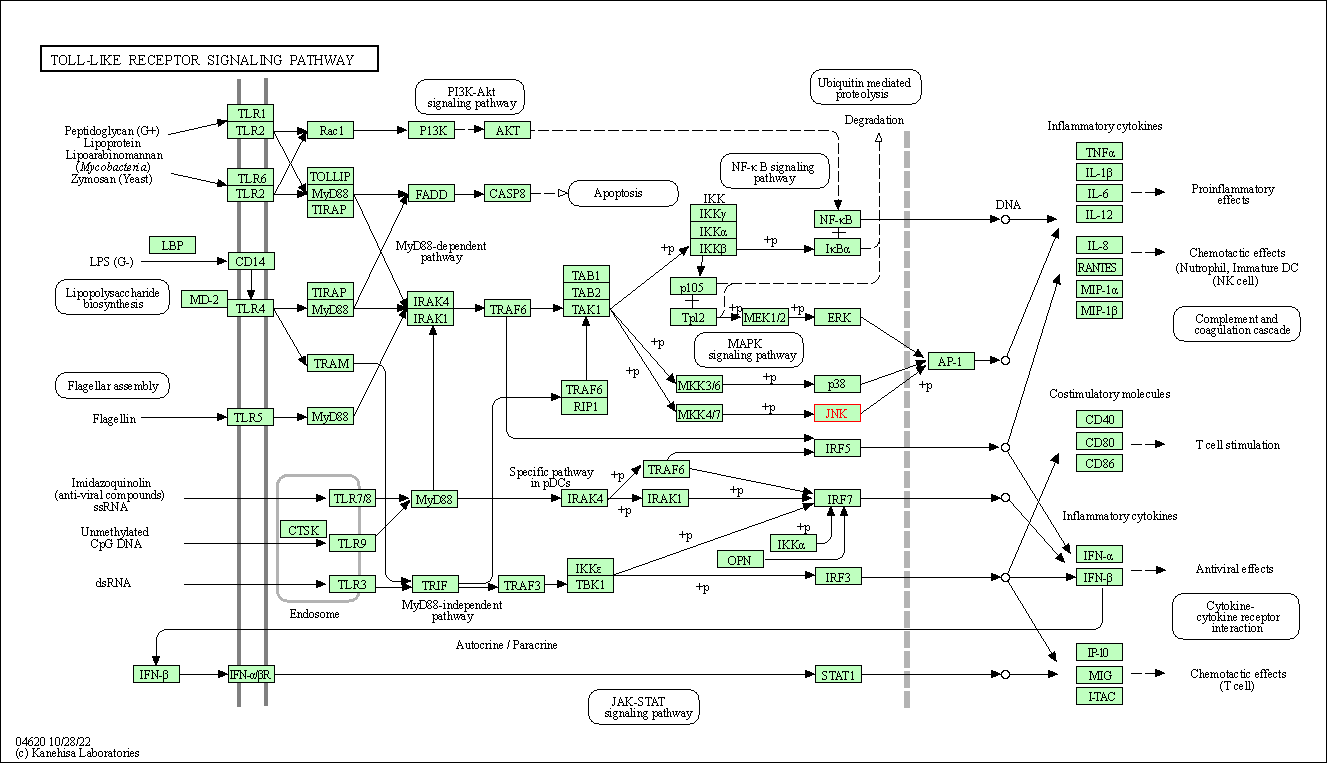
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| NOD-like receptor signaling pathway | hsa04621 | Affiliated Target |
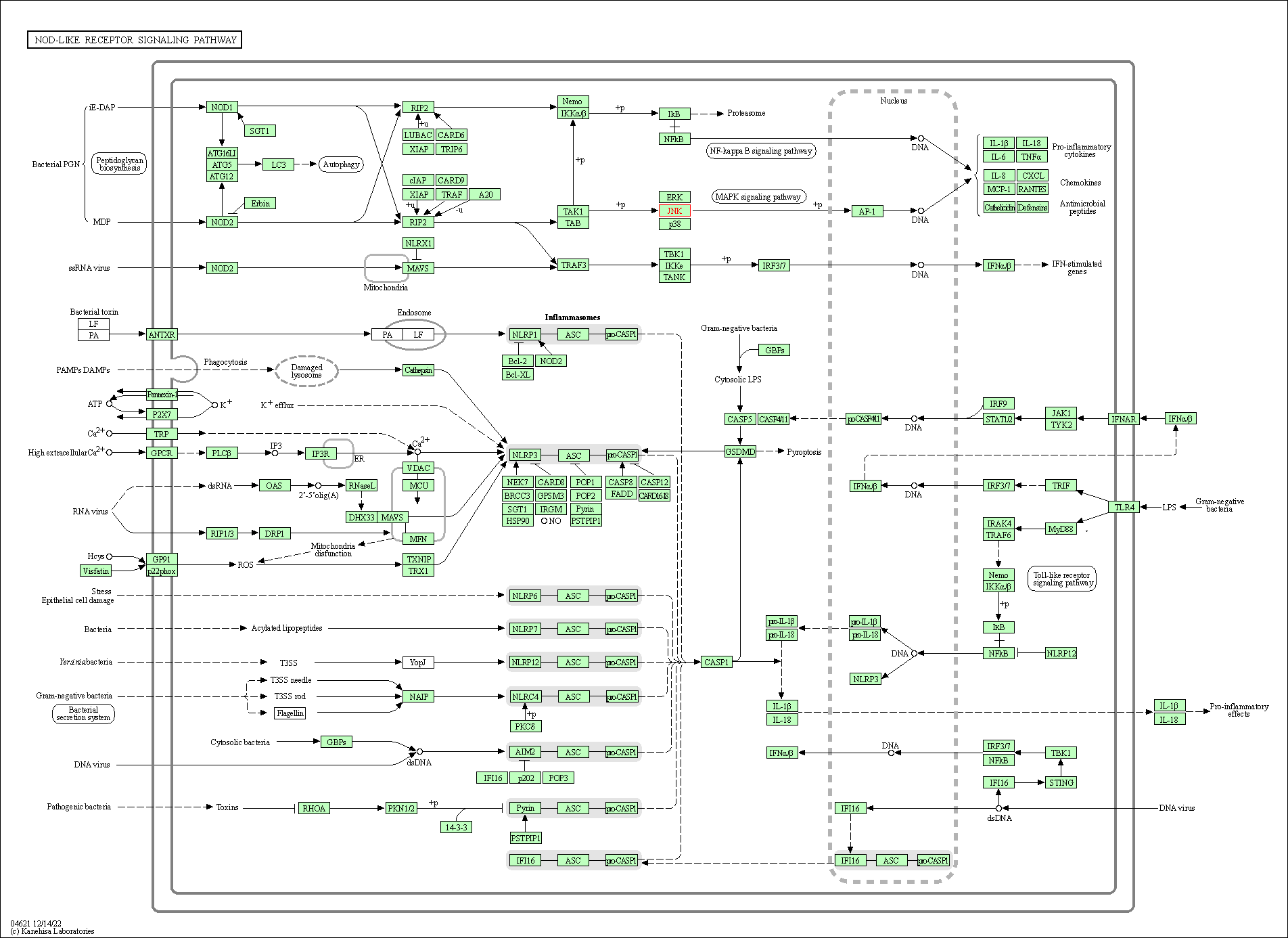
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| RIG-I-like receptor signaling pathway | hsa04622 | Affiliated Target |
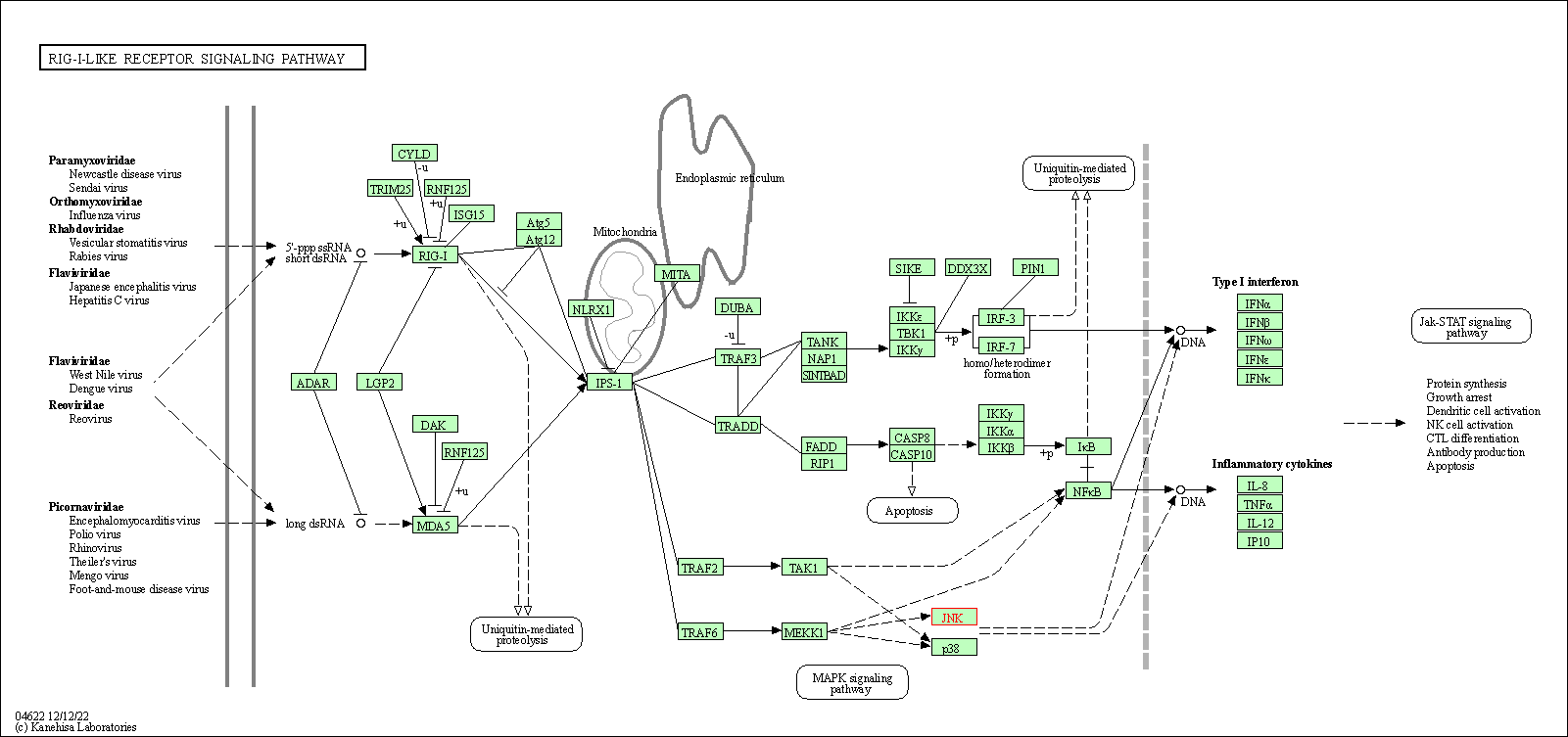
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| C-type lectin receptor signaling pathway | hsa04625 | Affiliated Target |
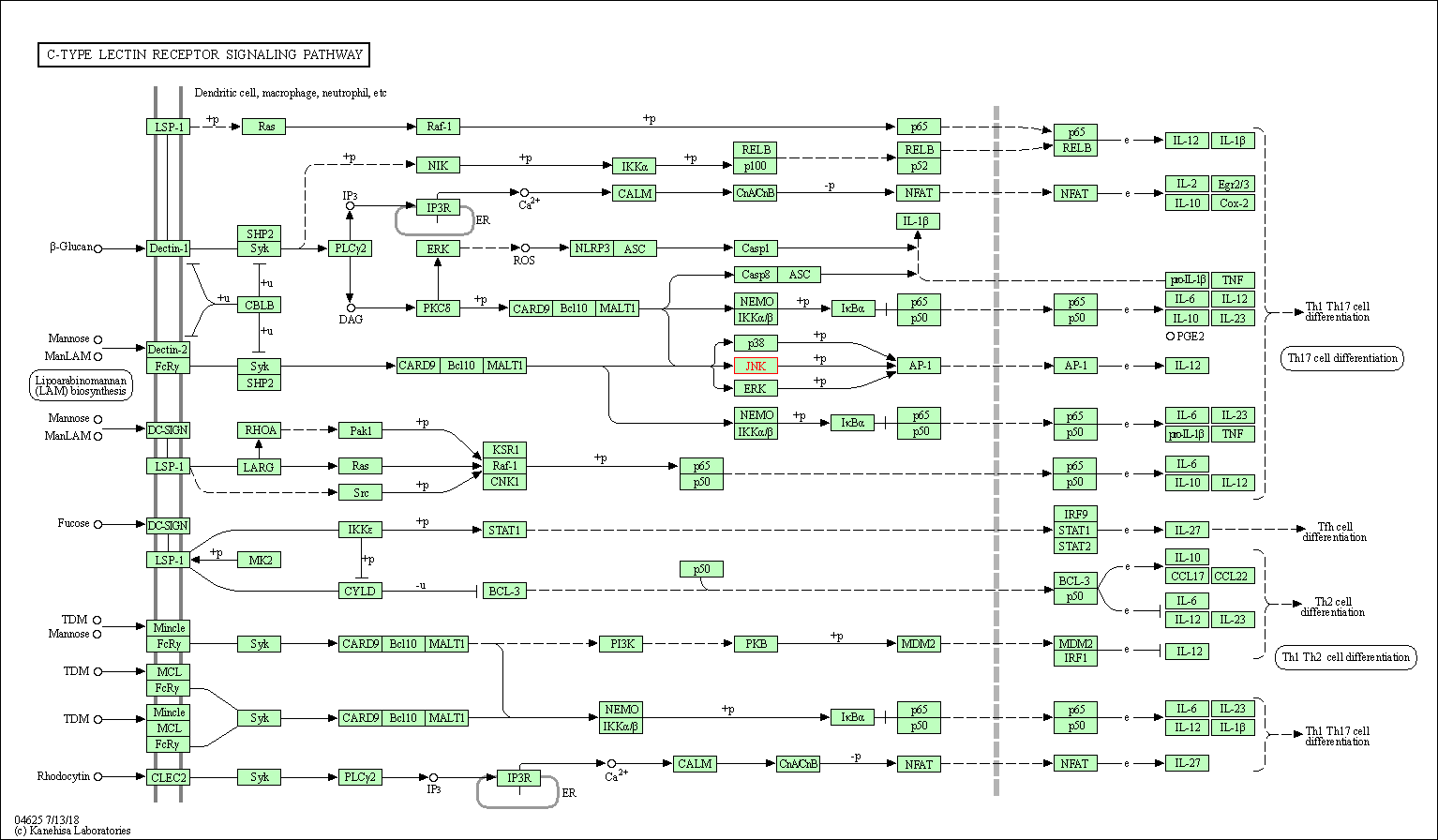
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| IL-17 signaling pathway | hsa04657 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th1 and Th2 cell differentiation | hsa04658 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| T cell receptor signaling pathway | hsa04660 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Fc epsilon RI signaling pathway | hsa04664 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| TNF signaling pathway | hsa04668 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Neurotrophin signaling pathway | hsa04722 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Prolactin signaling pathway | hsa04917 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Adipocytokine signaling pathway | hsa04920 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 9 | Degree centrality | 9.67E-04 | Betweenness centrality | 5.96E-05 |
|---|---|---|---|---|---|
| Closeness centrality | 2.29E-01 | Radiality | 1.40E+01 | Clustering coefficient | 1.94E-01 |
| Neighborhood connectivity | 4.08E+01 | Topological coefficient | 1.43E-01 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Drug Property Profile of Target | Top | |
|---|---|---|
| (1) Molecular Weight (mw) based Drug Clustering | (2) Octanol/Water Partition Coefficient (xlogp) based Drug Clustering | |
|
|
||
| (3) Hydrogen Bond Donor Count (hbonddonor) based Drug Clustering | (4) Hydrogen Bond Acceptor Count (hbondacc) based Drug Clustering | |
|
|
||
| (5) Rotatable Bond Count (rotbonds) based Drug Clustering | (6) Topological Polar Surface Area (polararea) based Drug Clustering | |
|
|
||
| "RO5" indicates the cutoff set by lipinski's rule of five; "D123AB" colored in GREEN denotes the no violation of any cutoff in lipinski's rule of five; "D123AB" colored in PURPLE refers to the violation of only one cutoff in lipinski's rule of five; "D123AB" colored in BLACK represents the violation of more than one cutoffs in lipinski's rule of five | ||
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| Target Regulators | Top | |||||
|---|---|---|---|---|---|---|
| Target-interacting Proteins | ||||||
| Target-Related Models and Studies | Top | |||||
|---|---|---|---|---|---|---|
| Target Validation | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | c-Jun N-terminal kinase inhibitors: a patent review (2010 - 2014).Expert Opin Ther Pat. 2015;25(8):849-72. | |||||
| REF 2 | How many drug targets are there Nat Rev Drug Discov. 2006 Dec;5(12):993-6. | |||||
| REF 3 | URL: http://www.guidetopharmacology.org Nucleic Acids Res. 2015 Oct 12. pii: gkv1037. The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. (Target id: 1498). | |||||
| REF 4 | Aminopyridine-based c-Jun N-terminal kinase inhibitors with cellular activity and minimal cross-kinase activity. J Med Chem. 2006 Jun 15;49(12):3563-80. | |||||
| REF 5 | The Protein Data Bank. Nucleic Acids Res. 2000 Jan 1;28(1):235-42. | |||||
| REF 6 | A systematic interaction map of validated kinase inhibitors with Ser/Thr kinases. Proc Natl Acad Sci U S A. 2007 Dec 18;104(51):20523-8. | |||||
| REF 7 | Discovery of potent and selective covalent inhibitors of JNK. Chem Biol. 2012 Jan 27;19(1):140-54. | |||||
| REF 8 | Synthesis and preclinical evaluations of 2-(2-fluorophenyl)-6,7-methylenedioxyquinolin-4-one monosodium phosphate (CHM-1-P-Na) as a potent antitumo... J Med Chem. 2010 Feb 25;53(4):1616-26. | |||||
| REF 9 | Design and synthesis of 6-anilinoindazoles as selective inhibitors of c-Jun N-terminal kinase-3. Bioorg Med Chem Lett. 2005 Nov 15;15(22):5095-9. | |||||
| REF 10 | The crystal structure of AMP-bound JNK3 | |||||
| REF 11 | Discovery of CC-930, an orally active anti-fibrotic JNK inhibitor. Bioorg Med Chem Lett. 2012 Feb 1;22(3):1433-8. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

