Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T00645
|
|||||
| Target Name |
HUMAN protein kinase cAMP-activated catalytic subunit alpha (PRKACA)
|
|||||
| Synonyms |
PKA C-alpha; cAMP-dependent protein kinase catalytic subunit alpha
Click to Show/Hide
|
|||||
| Gene Name |
PRKACA
|
|||||
| Function |
Human protein protein kinase cAMP-activated catalytic subunit alpha interacts with SARS-CoV-2 Nsp13 protein with high significance, which indicates PRKACA as a potential therapeutic target.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.11
|
|||||
| Sequence |
MGNAAAAKKGSEQESVKEFLAKAKEDFLKKWESPAQNTAHLDQFERIKTLGTGSFGRVML
VKHKETGNHYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVKLEFSFKDNSNLYMV MEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDQQGY IQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFF ADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVNDIKNHKWFAT TDWIAIYQRKVEAPFIPKFKGPGDTSNFDDYEEEEIRVSINEKCGKEFSEF Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | AlphaFold | ||||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Adenosine triphosphate | Ligand Info | |||||
| Structure Description | Crystal structure of human cAMP-dependent protein kinase A (catalytic alpha subunit), exon 1 deletion | PDB:4WB8 | ||||
| Method | X-ray diffraction | Resolution | 1.55 Å | Mutation | No | [2] |
| PDB Sequence |
GVKEFLAKAK
23 EDFLKKWESP33 AQNTAHLDQF43 ERIKTLGTGS53 FGRVMLVKHK63 ETGNHYAMKI 73 LDKQKVVKLK83 QIEHTLNEKR93 ILQAVNFPFL103 VKLEFSFKDN113 SNLYMVMEYV 123 PGGEMFSHLR133 RIGRFEPHAR144 FYAAQIVLTF154 EYLHSLDLIY164 RDLKPENLLI 174 DQQGYIQVTD184 FGFAKRVKGR194 TWLCGTPEYL205 APEIILSKGY215 NKAVDWWALG 225 VLIYEMAAGY235 PPFFADQPIQ245 IYEKIVSGKV255 RFPSHFSSDL265 KDLLRNLLQV 275 DLTKRFGNLK285 NGVNDIKNHK295 WFATTDWIAI305 YQRKVEAPFI315 PKFKGPGDTS 325 NFDDYEEEEI335 RVINEKCGKE346 FSEF
|
|||||
|
|
LEU49
3.921
GLY50
3.561
THR51
3.810
GLY52
3.352
SER53
2.769
PHE54
2.904
GLY55
2.840
VAL57
3.236
ALA70
3.422
LYS72
2.823
GLU91
4.846
VAL104
3.582
MET120
3.483
|
|||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: AT9283 | Ligand Info | |||||
| Structure Description | Protein kinase A mutants as surrogate model for Aurora B with AT9283 inhibitor | PDB:5N23 | ||||
| Method | X-ray diffraction | Resolution | 2.09 Å | Mutation | Yes | [3] |
| PDB Sequence |
KGEQESVKEF
18 LAKAKEDFLK28 KWESPAQNTA38 HLDQFERIRT48 LGTGSFGRVM58 LVKHKETGNH 68 YAMKILDKQK78 VVKLKQIEHT88 LNEKRIQQAV98 NFPFLVKLEF108 SFKDNSNLYM 118 VLEYAPGGEM128 FSHLRRIGRF138 EPHARFYAAQ149 IVLTFEYLHS159 LDLIYRDLKP 169 ENLLIDQQGY179 IKVADFGFAK189 RVKGRTWLCG200 TPEYLAPEII210 LSKGYNKAVD 220 WWALGVLIYE230 MAAGYPPFFA240 DQPIQIYEKI250 VSGKVRFPSH260 FSSDLKDLLR 270 NLLQVDLTKR280 FGNLKNGVND290 IKNHKWFATT300 DWIAIYQRKV310 EAPFIPKFSN 326 FDDYEEEEIR336 VINEKCGKEF347 SEF
|
|||||
|
|
LEU49
3.670
GLY50
3.572
THR51
3.880
VAL57
3.928
ALA70
3.416
VAL104
4.737
LEU120
3.668
GLU121
2.736
TYR122
3.380
ALA123
2.746
PRO124
3.600
|
|||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| MAPK signaling pathway | hsa04010 | Affiliated Target |
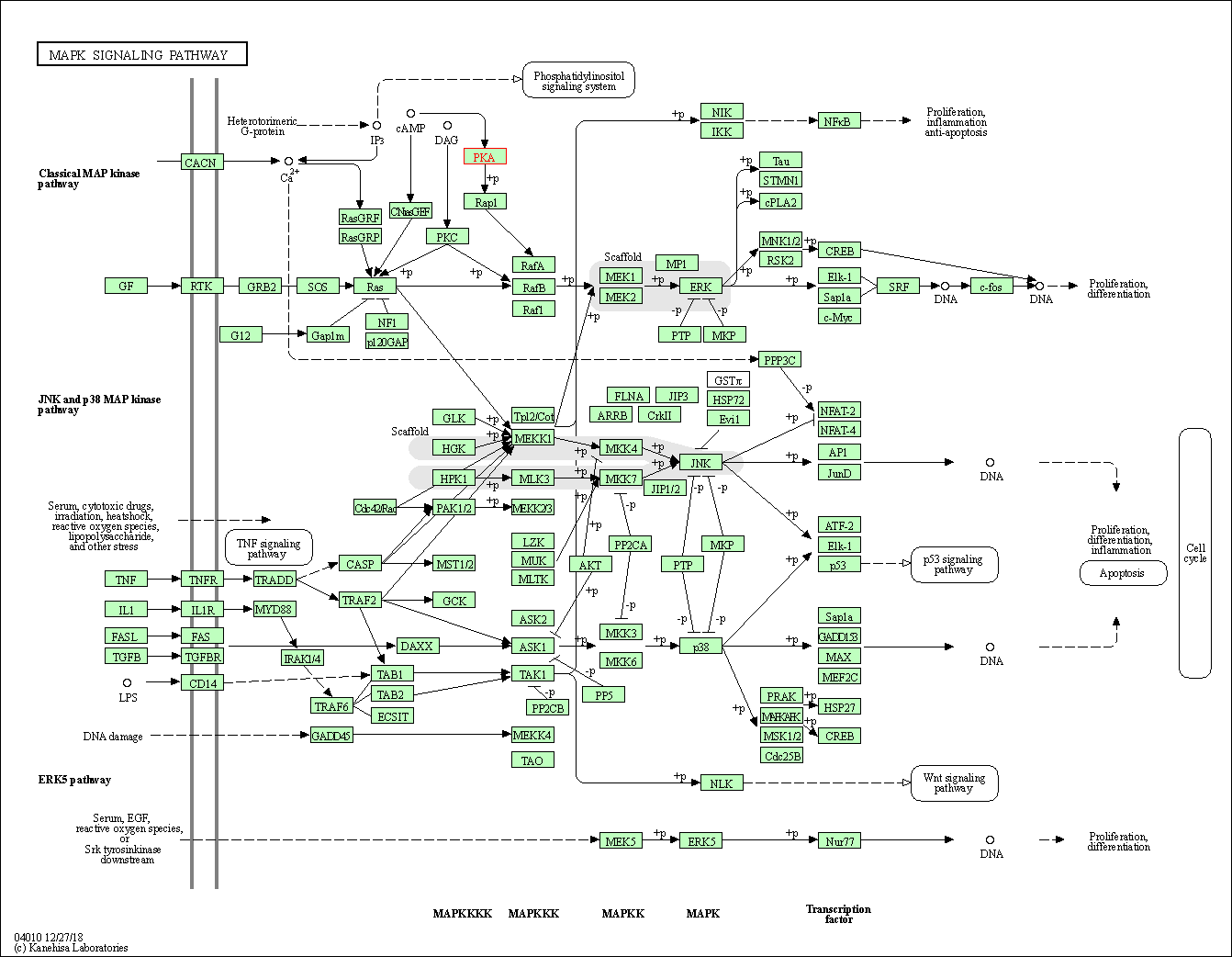
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Ras signaling pathway | hsa04014 | Affiliated Target |
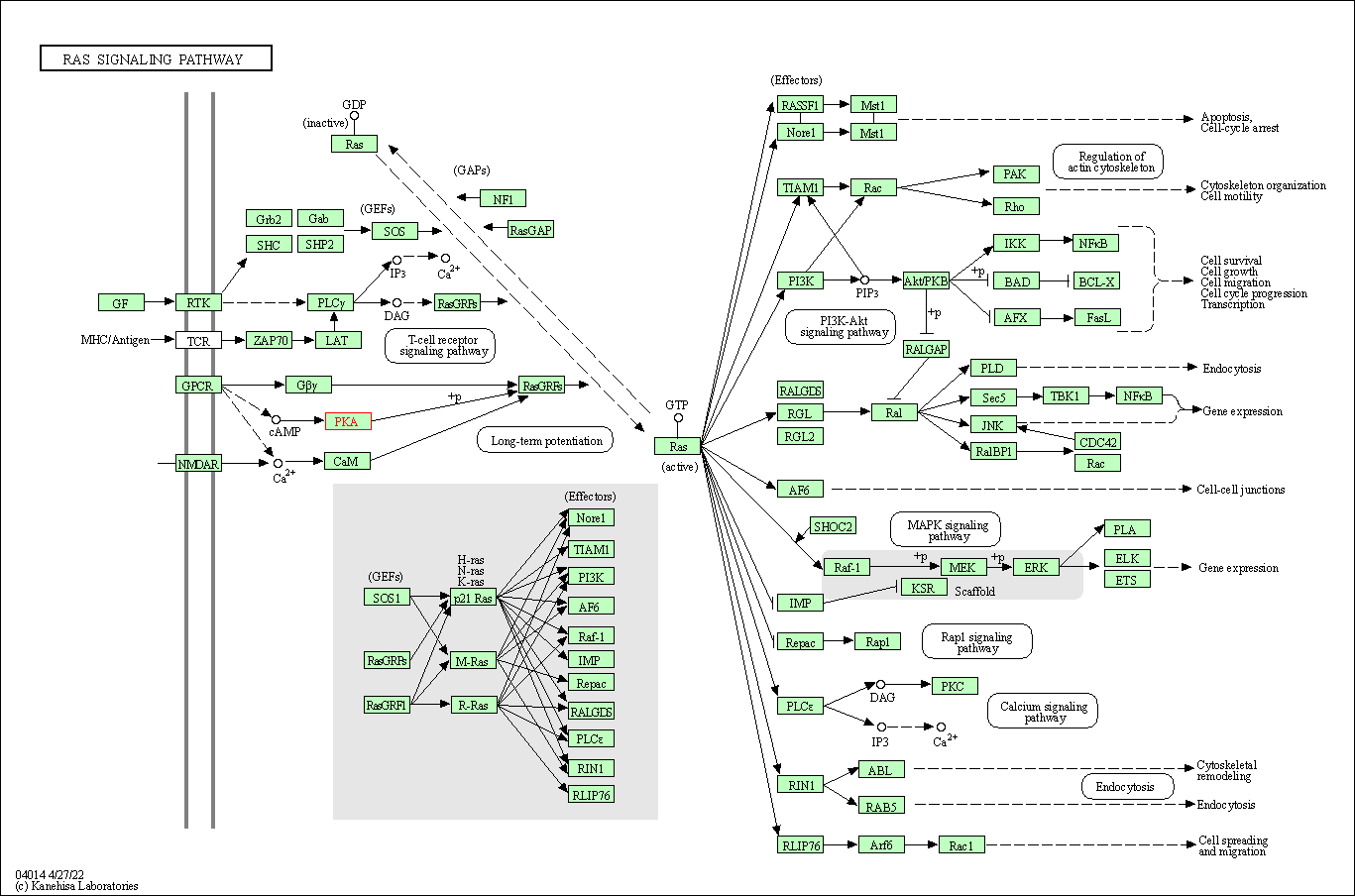
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Calcium signaling pathway | hsa04020 | Affiliated Target |
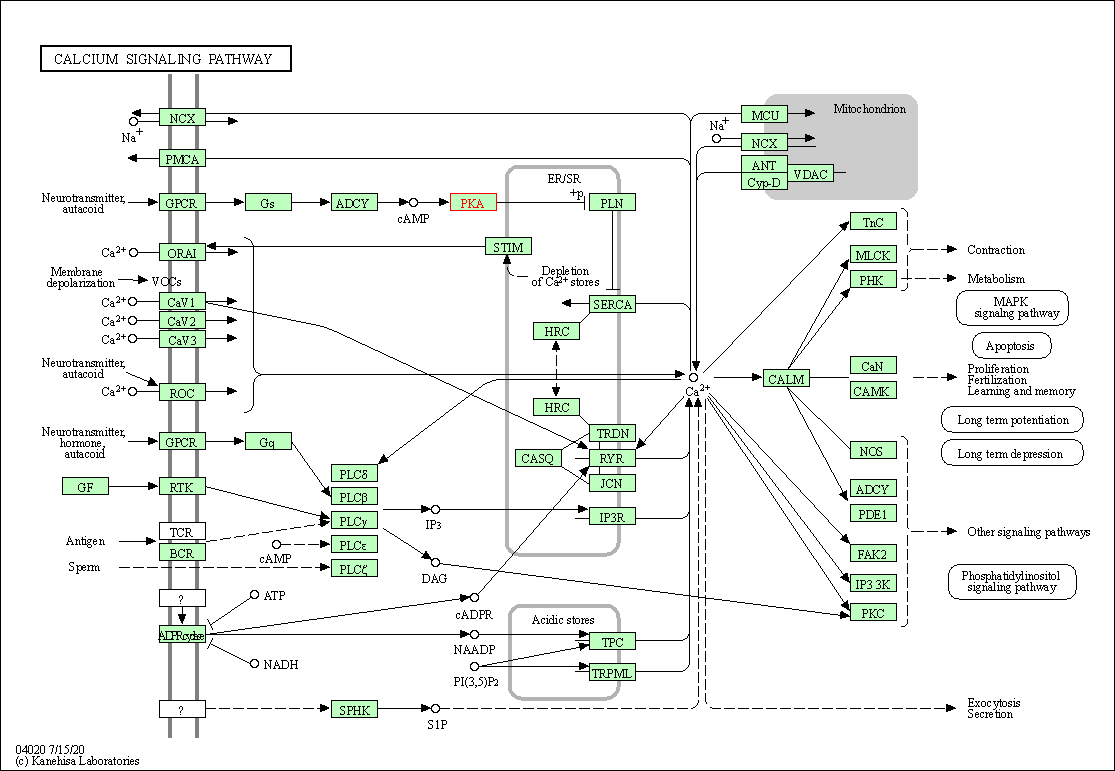
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| cAMP signaling pathway | hsa04024 | Affiliated Target |
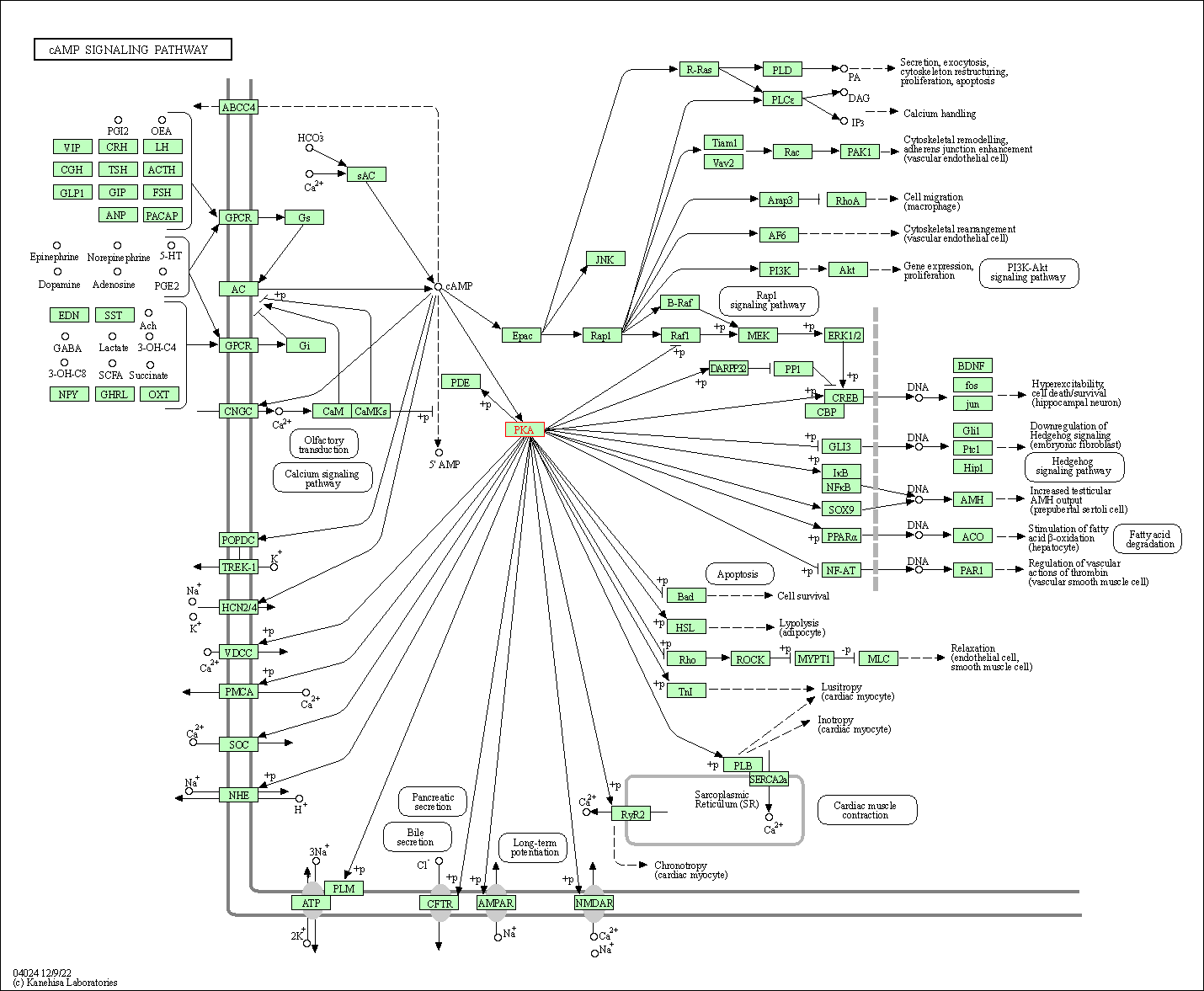
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Chemokine signaling pathway | hsa04062 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Oocyte meiosis | hsa04114 | Affiliated Target |
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Longevity regulating pathway | hsa04211 | Affiliated Target |
|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |
|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Adrenergic signaling in cardiomyocytes | hsa04261 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Vascular smooth muscle contraction | hsa04270 | Affiliated Target |
|
| Class: Organismal Systems => Circulatory system | Pathway Hierarchy | ||
| Wnt signaling pathway | hsa04310 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Hedgehog signaling pathway | hsa04340 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Tight junction | hsa04530 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Gap junction | hsa04540 | Affiliated Target |
|
| Class: Cellular Processes => Cellular community - eukaryotes | Pathway Hierarchy | ||
| Platelet activation | hsa04611 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Circadian entrainment | hsa04713 | Affiliated Target |
|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Thermogenesis | hsa04714 | Affiliated Target |
|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Long-term potentiation | hsa04720 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Retrograde endocannabinoid signaling | hsa04723 | Affiliated Target |
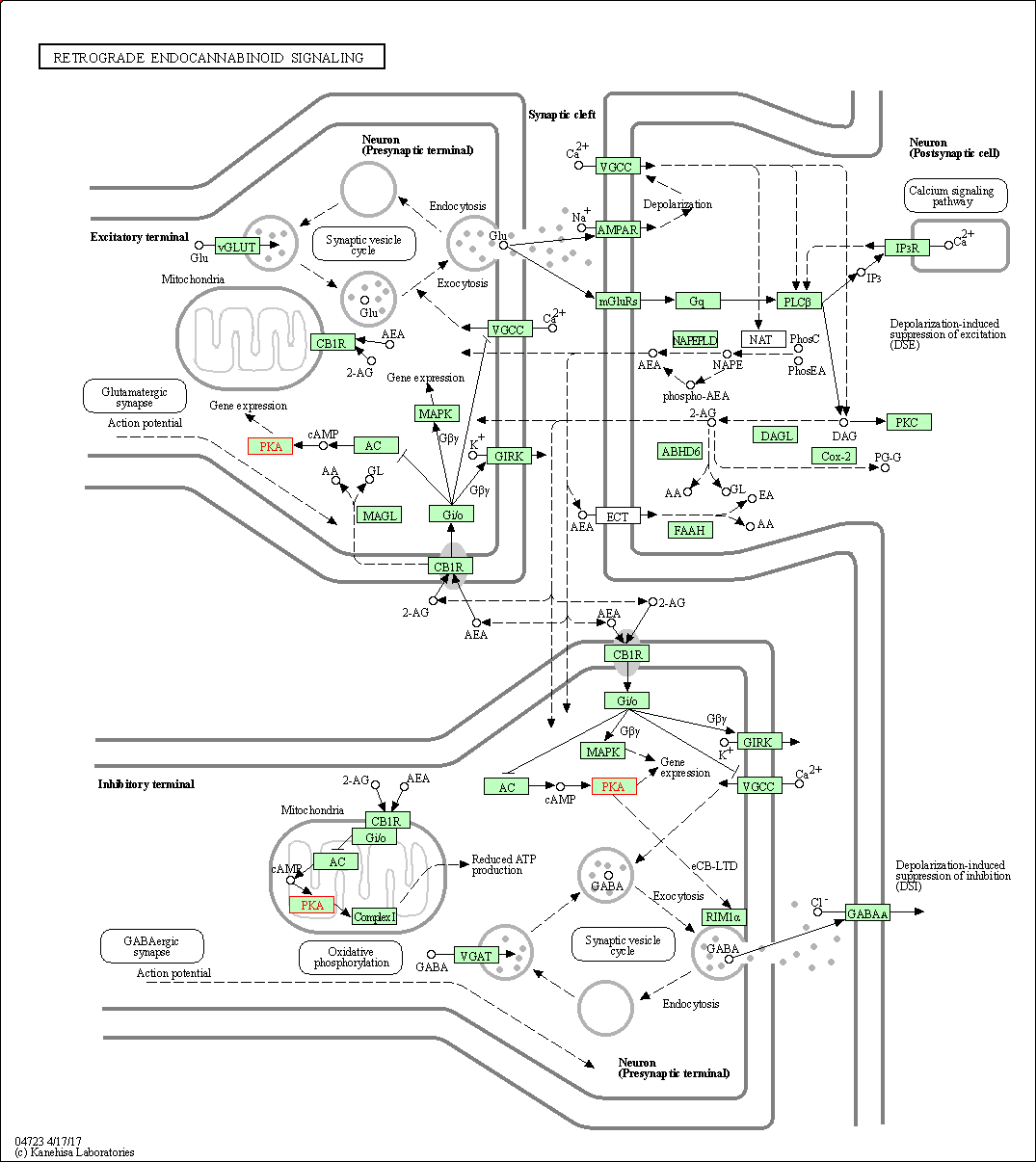
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Glutamatergic synapse | hsa04724 | Affiliated Target |
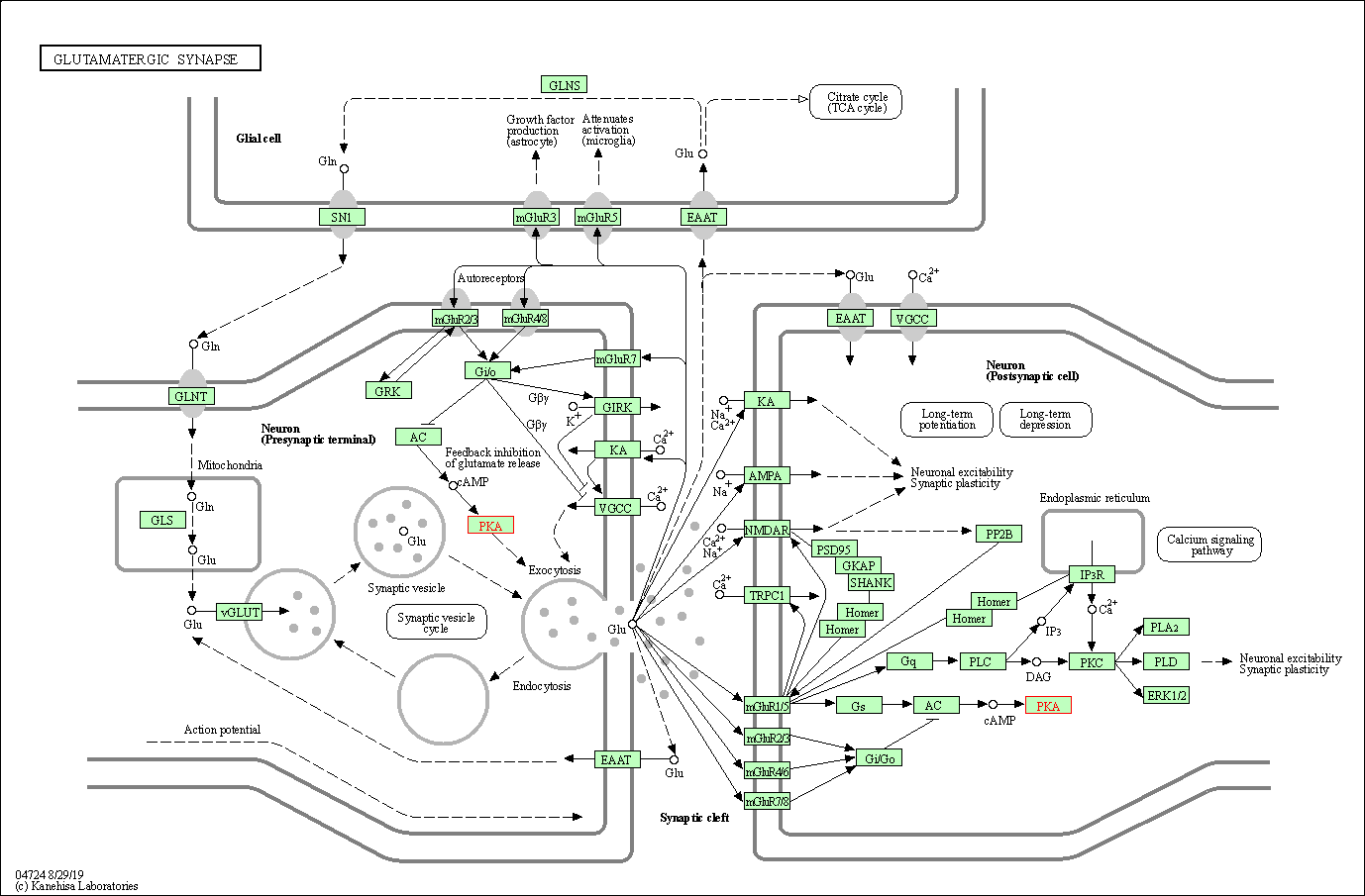
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Cholinergic synapse | hsa04725 | Affiliated Target |
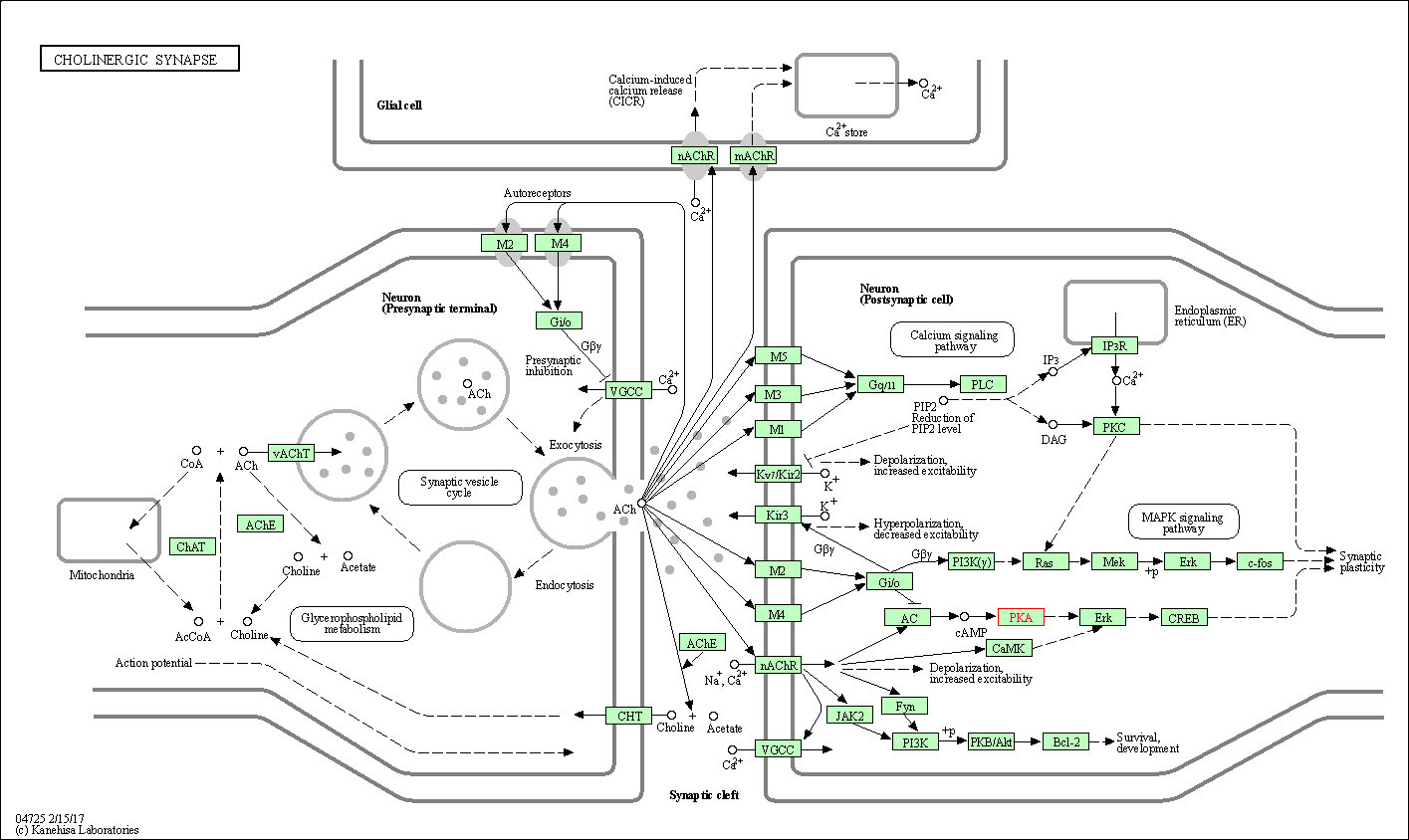
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Serotonergic synapse | hsa04726 | Affiliated Target |
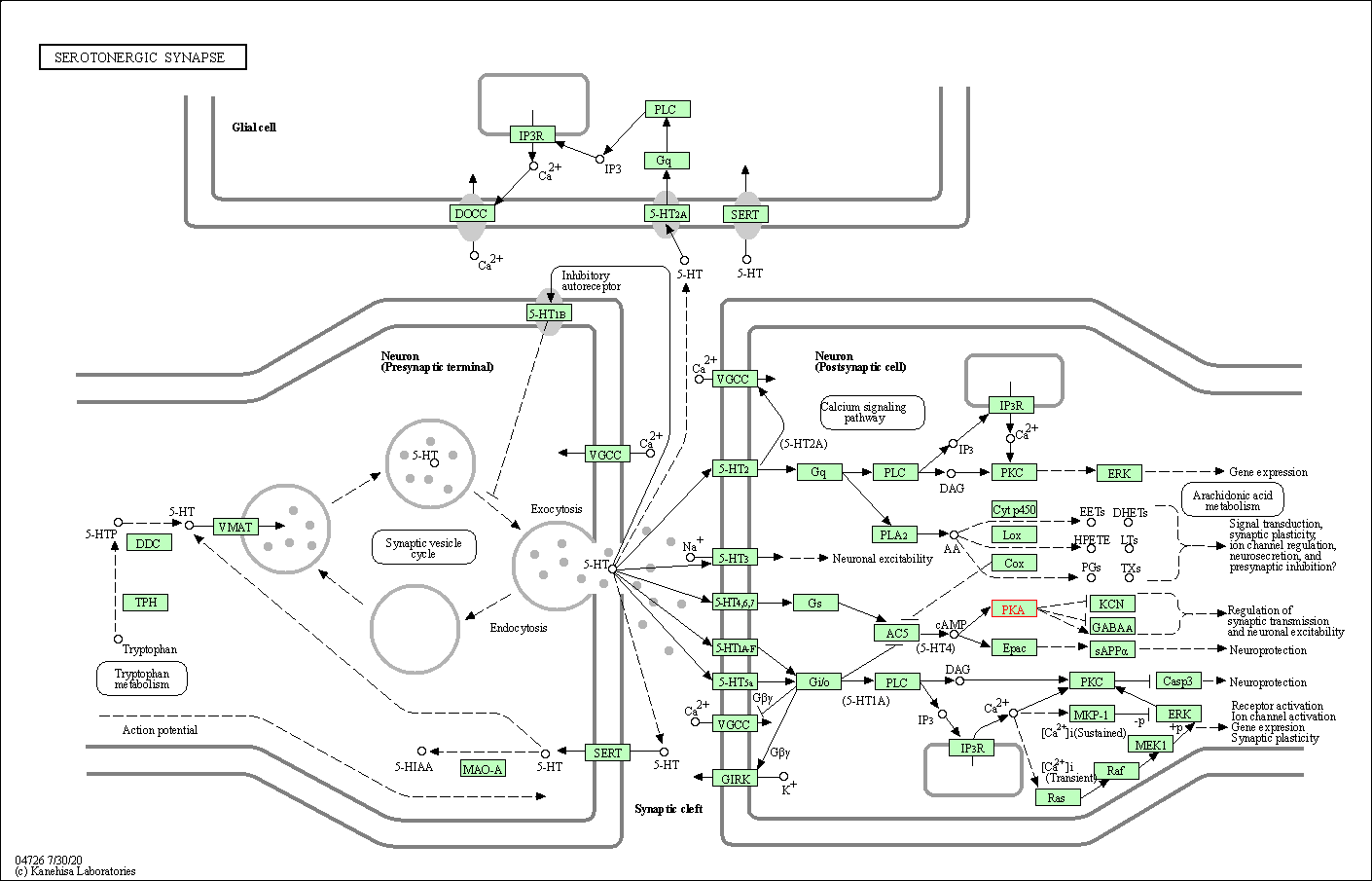
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| GABAergic synapse | hsa04727 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Dopaminergic synapse | hsa04728 | Affiliated Target |
|
| Class: Organismal Systems => Nervous system | Pathway Hierarchy | ||
| Olfactory transduction | hsa04740 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Taste transduction | hsa04742 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Inflammatory mediator regulation of TRP channels | hsa04750 | Affiliated Target |
|
| Class: Organismal Systems => Sensory system | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Insulin secretion | hsa04911 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| GnRH signaling pathway | hsa04912 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Ovarian steroidogenesis | hsa04913 | Affiliated Target |
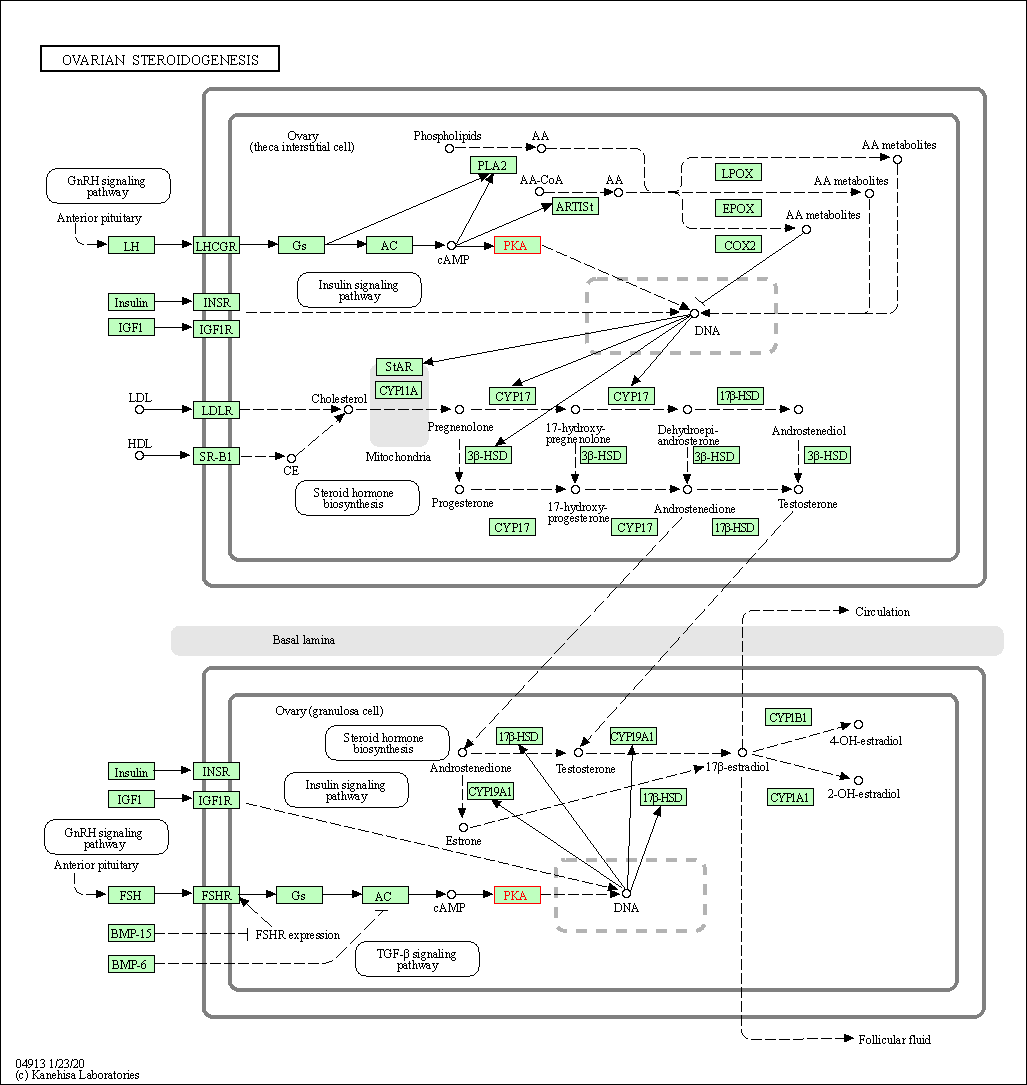
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Progesterone-mediated oocyte maturation | hsa04914 | Affiliated Target |
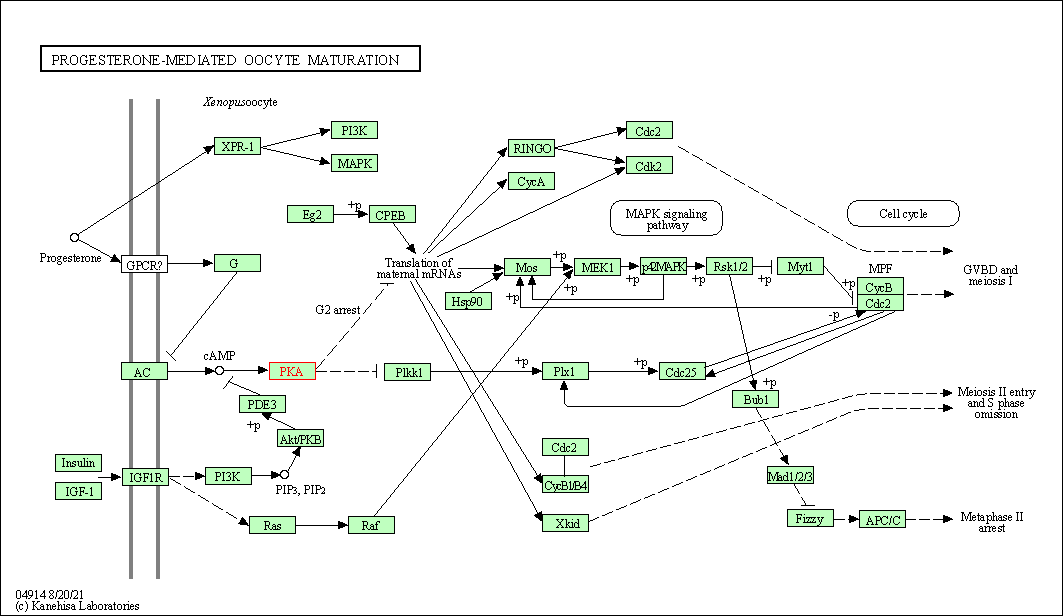
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Estrogen signaling pathway | hsa04915 | Affiliated Target |
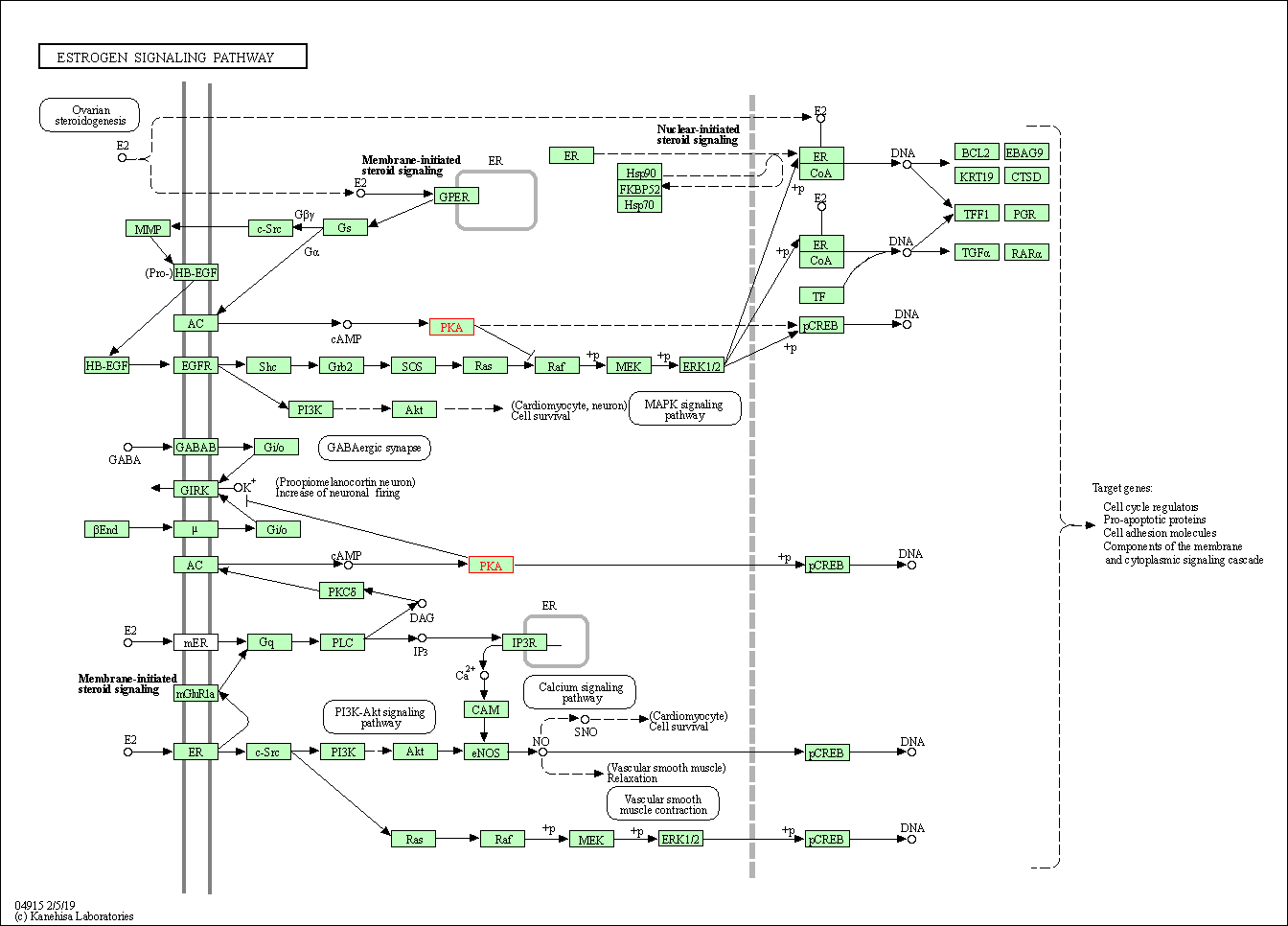
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Melanogenesis | hsa04916 | Affiliated Target |
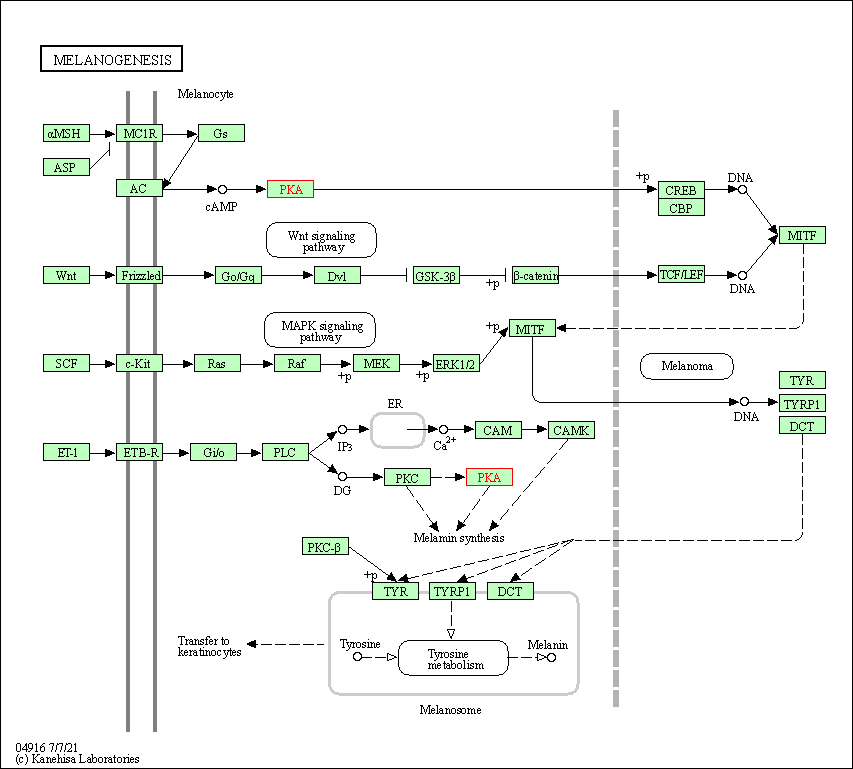
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone synthesis | hsa04918 | Affiliated Target |
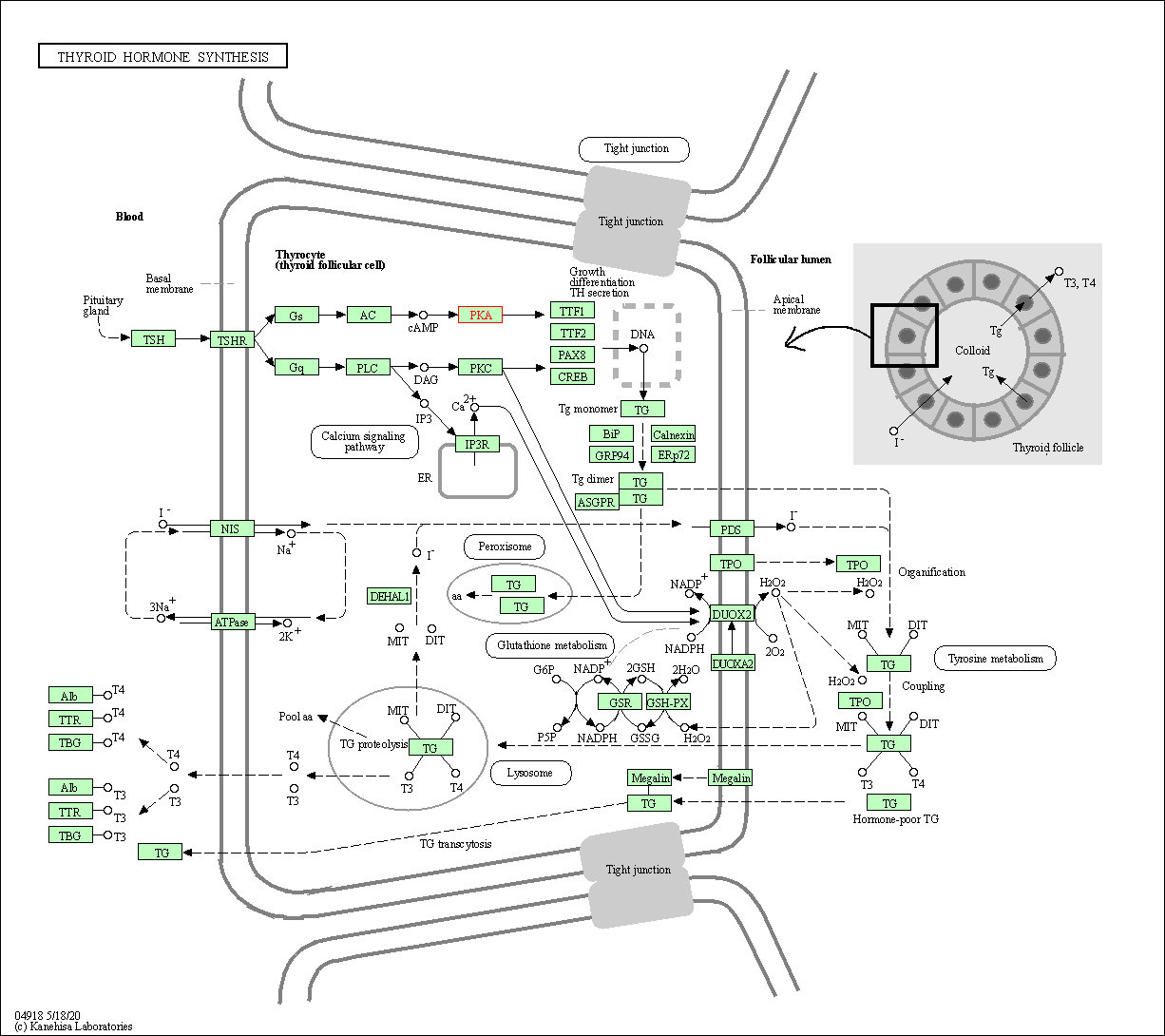
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |
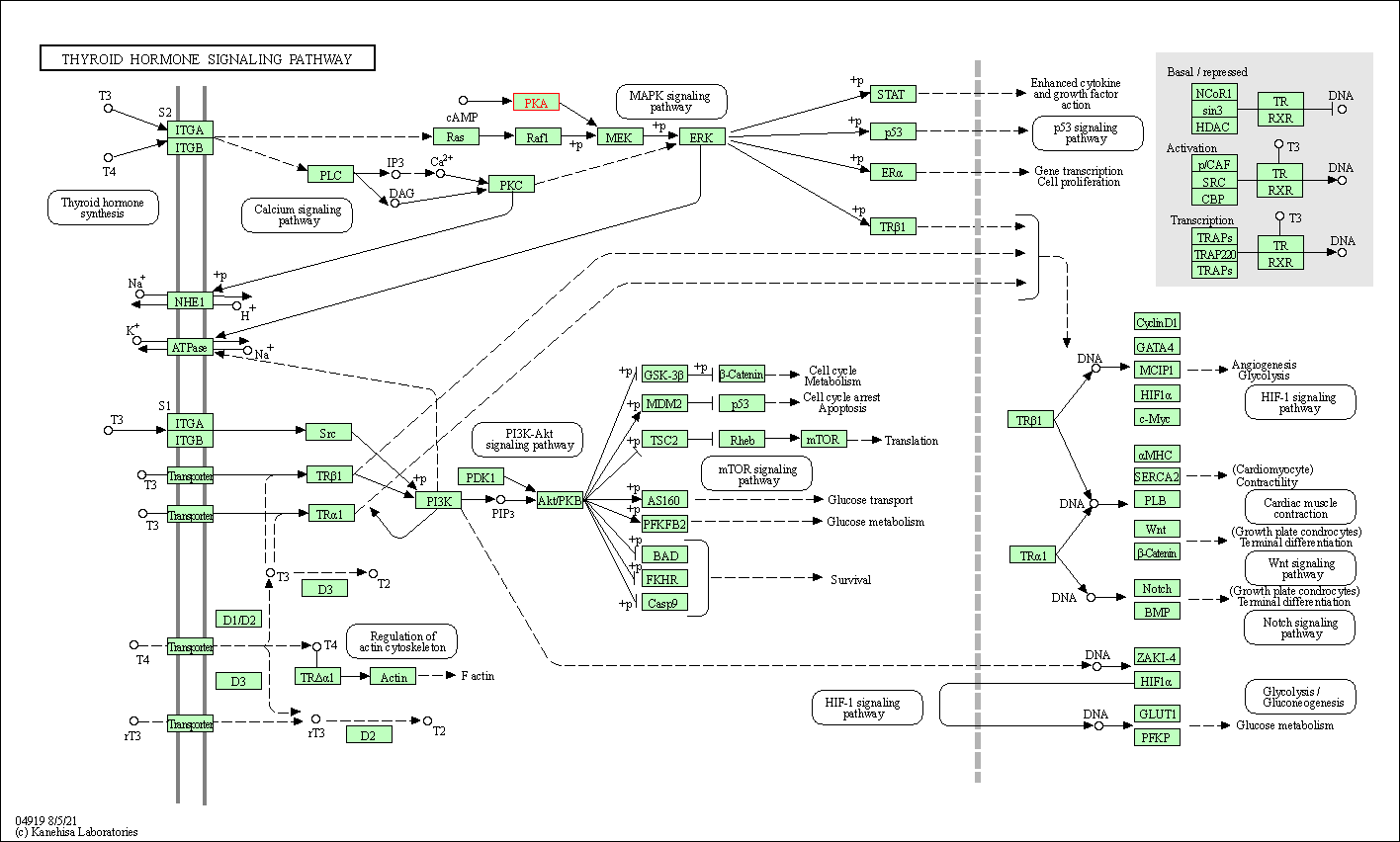
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Oxytocin signaling pathway | hsa04921 | Affiliated Target |
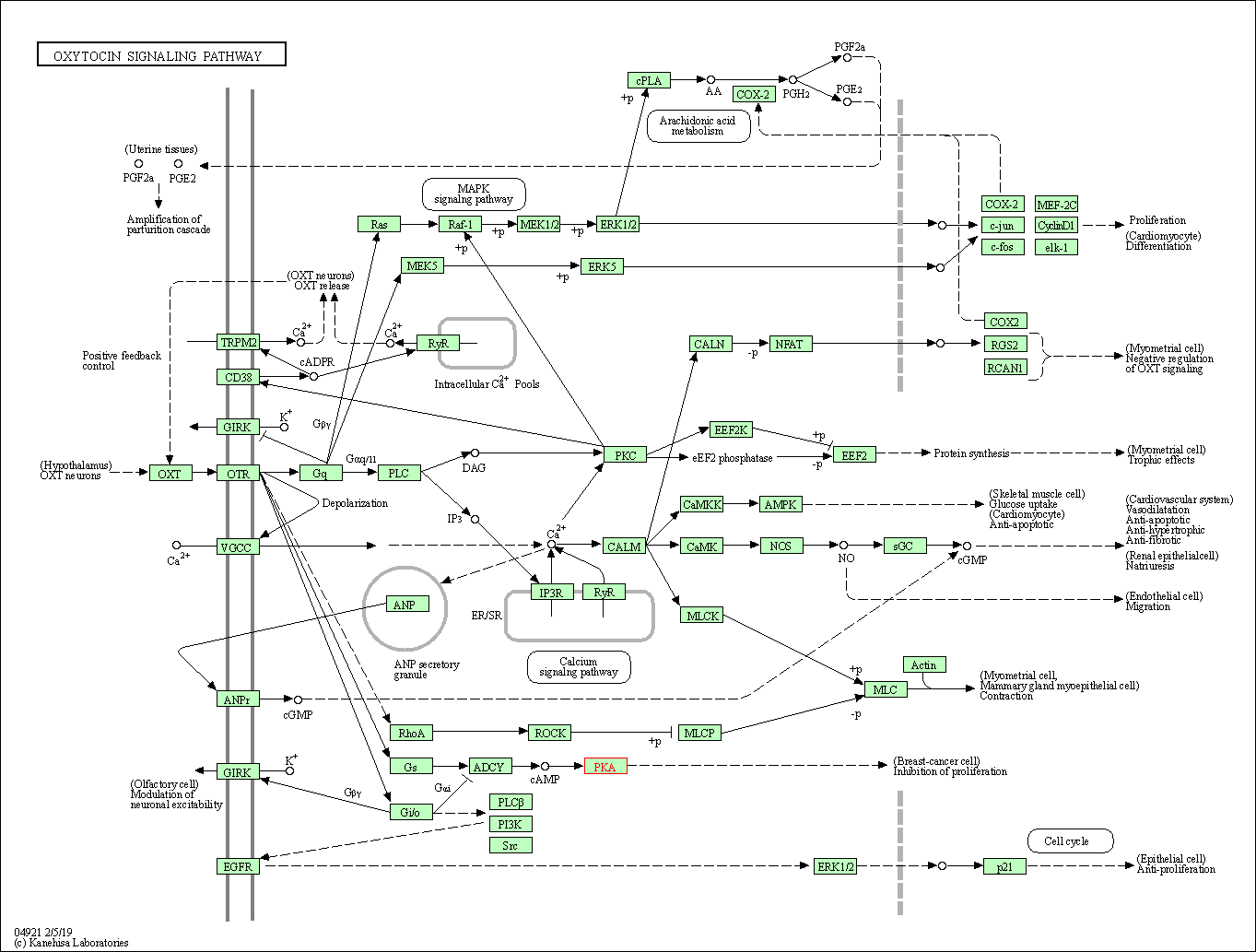
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Glucagon signaling pathway | hsa04922 | Affiliated Target |
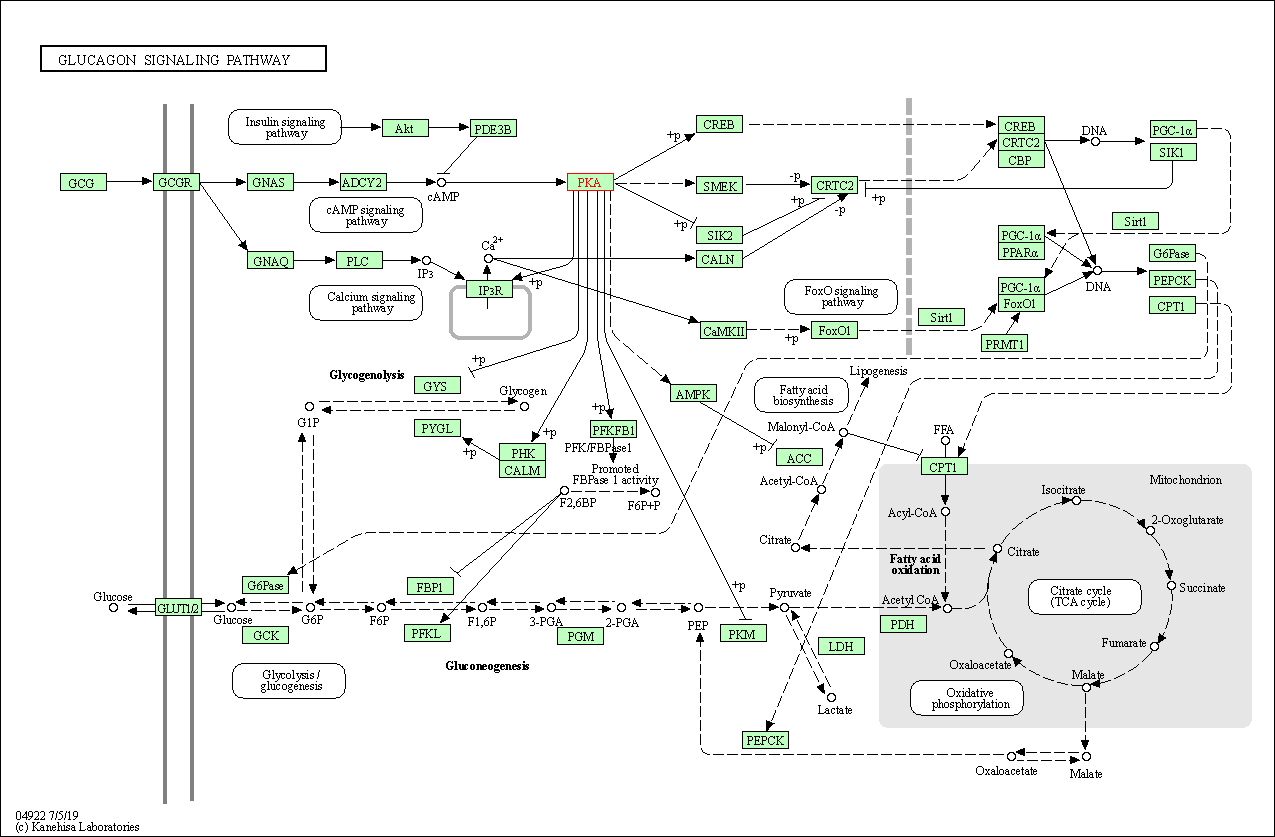
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Regulation of lipolysis in adipocytes | hsa04923 | Affiliated Target |
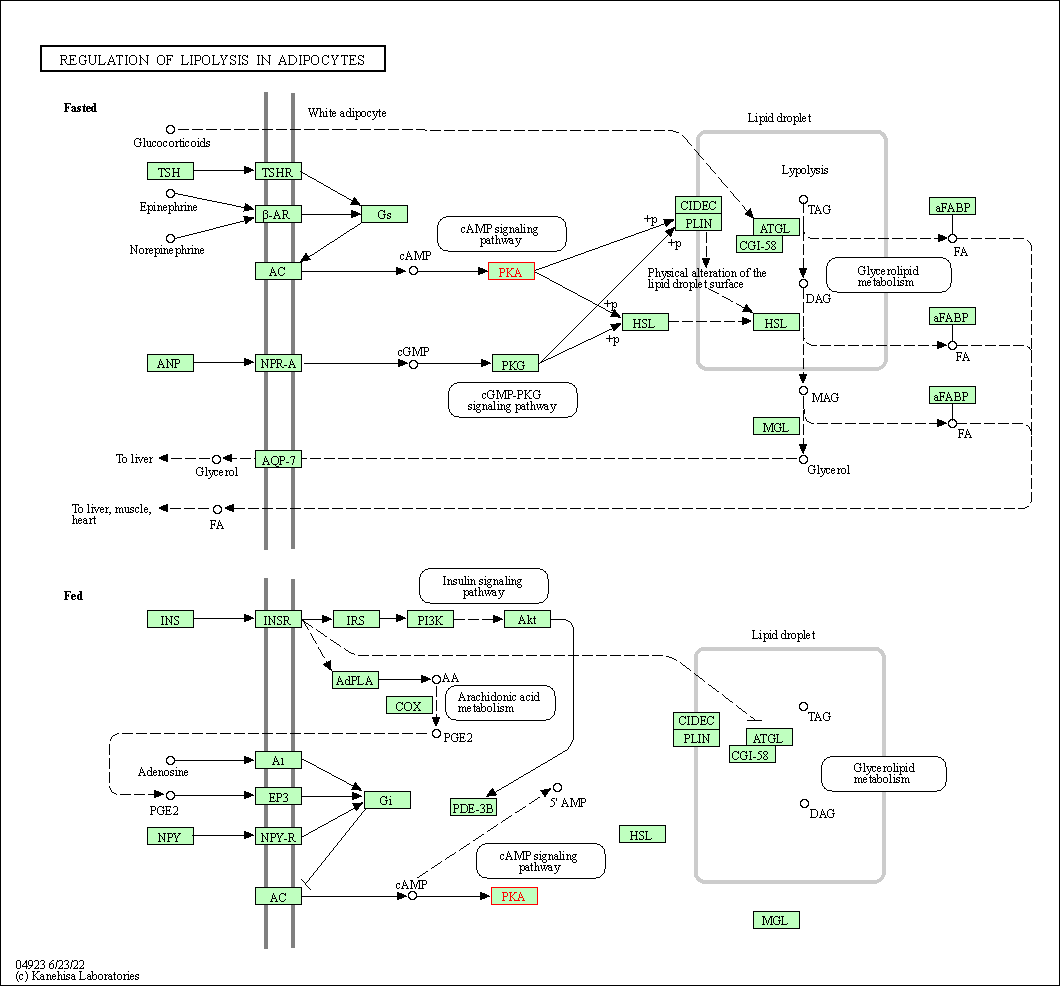
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Renin secretion | hsa04924 | Affiliated Target |
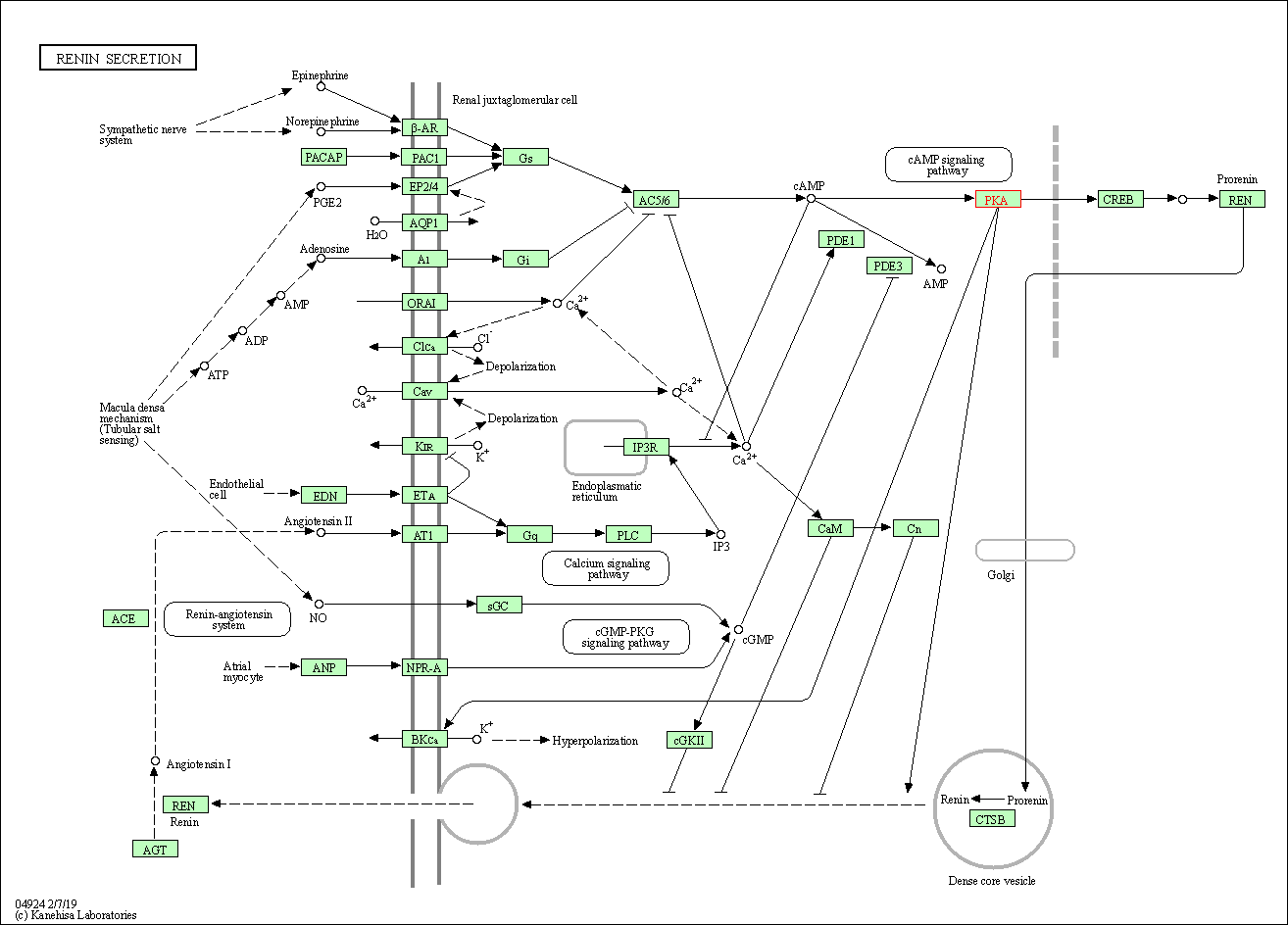
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Aldosterone synthesis and secretion | hsa04925 | Affiliated Target |
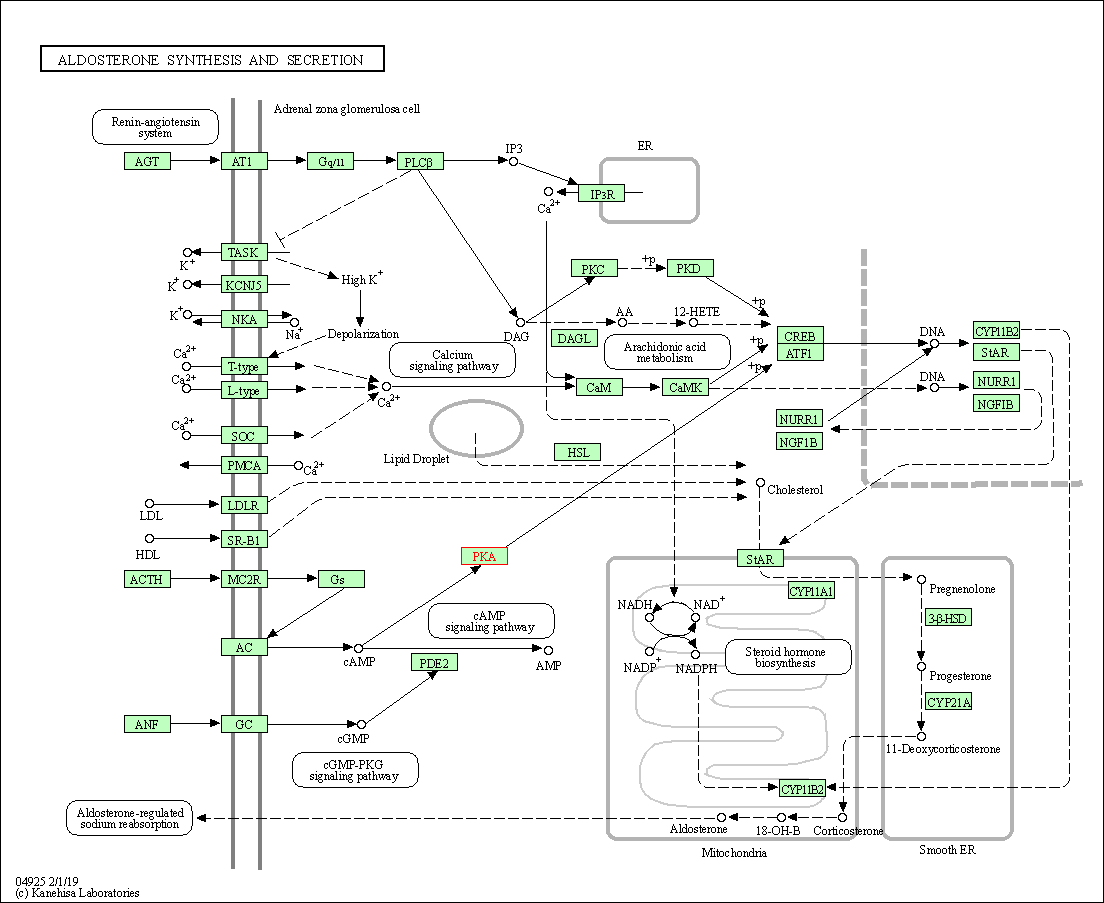
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Relaxin signaling pathway | hsa04926 | Affiliated Target |
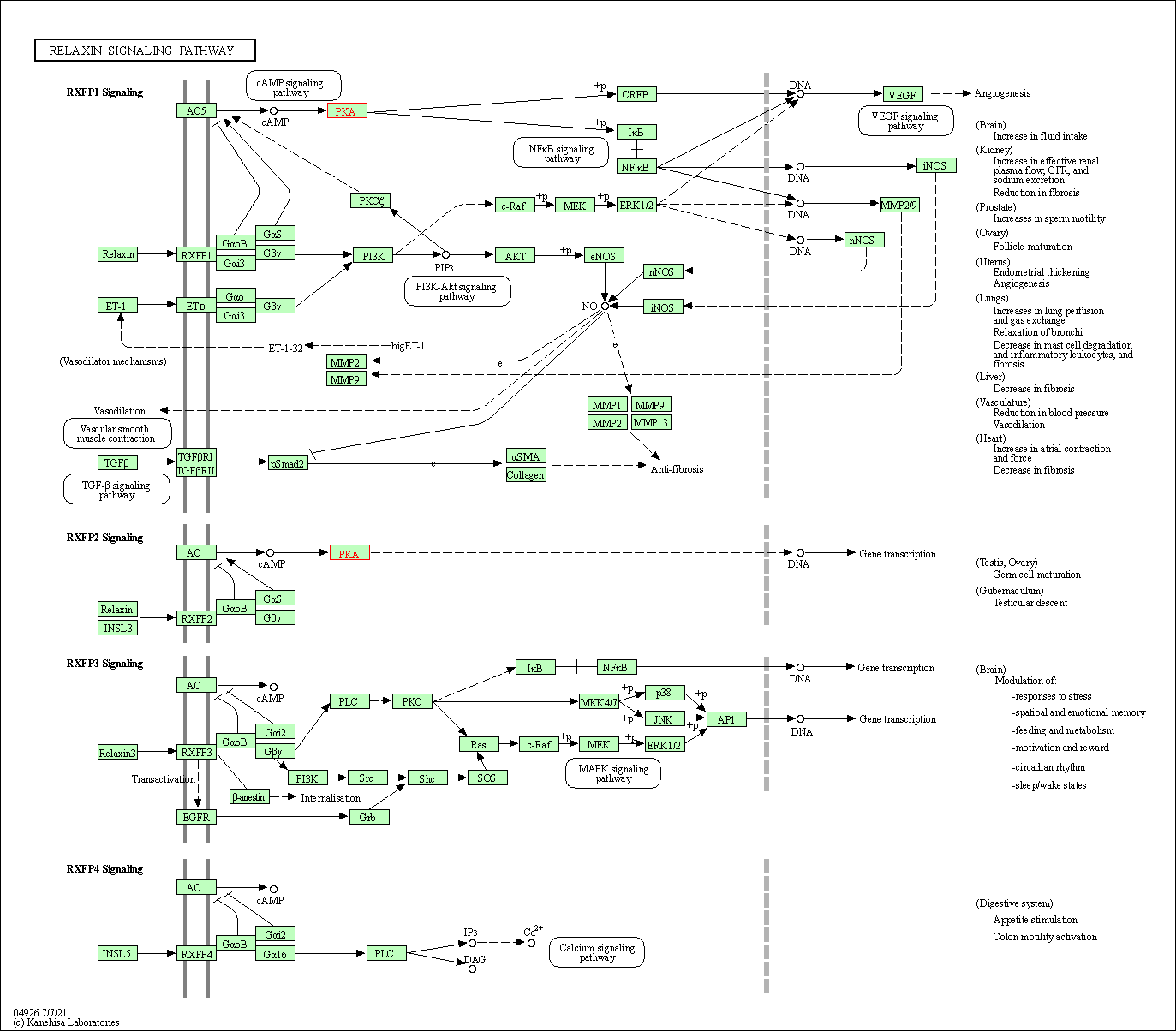
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Cortisol synthesis and secretion | hsa04927 | Affiliated Target |
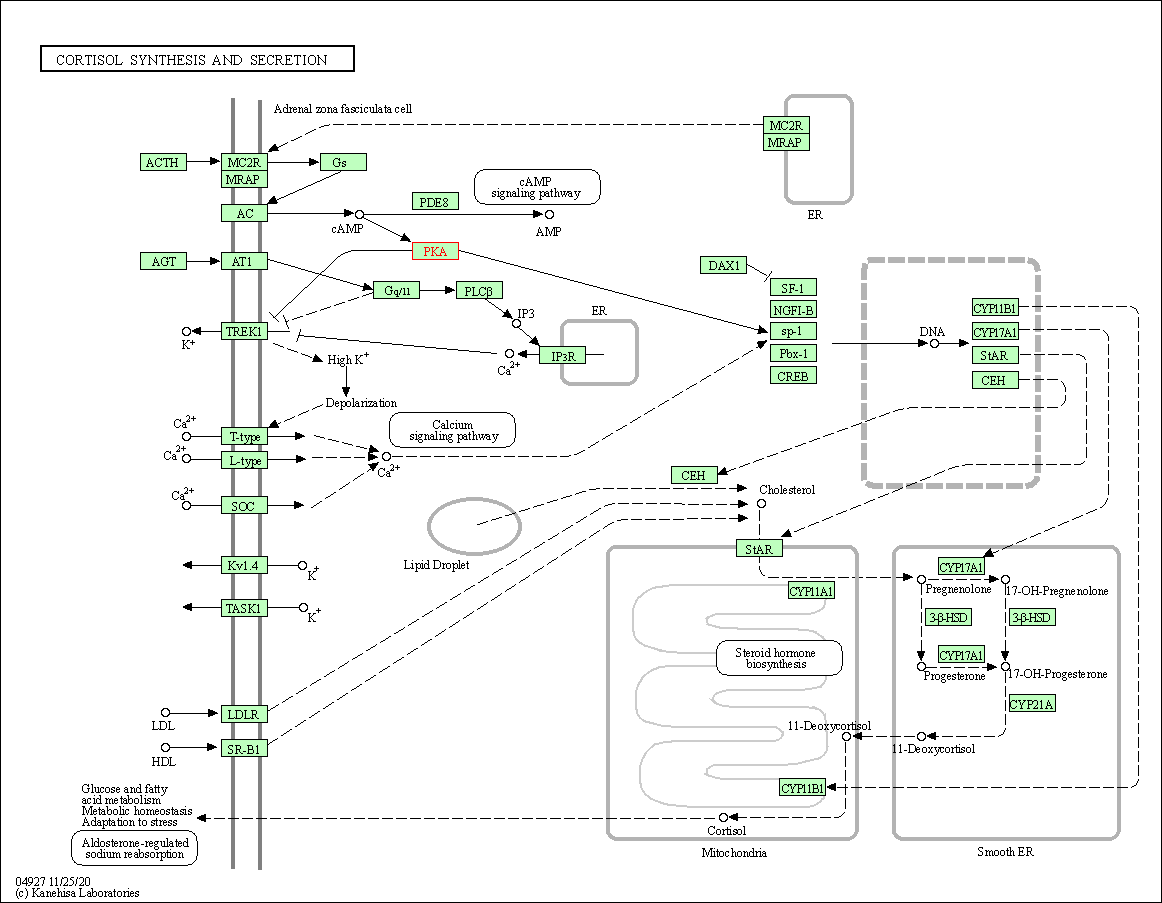
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Parathyroid hormone synthesis, secretion and action | hsa04928 | Affiliated Target |
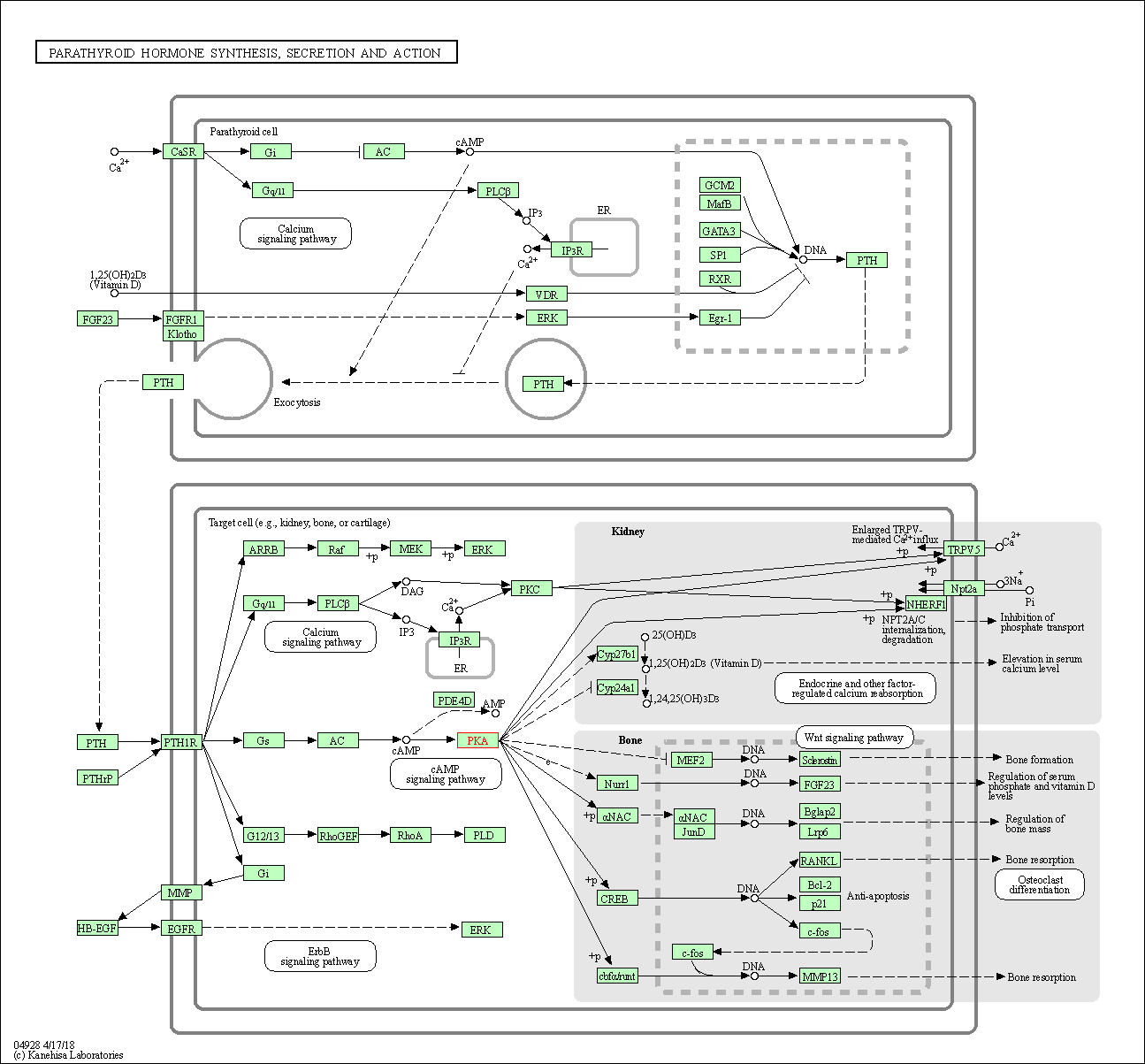
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |
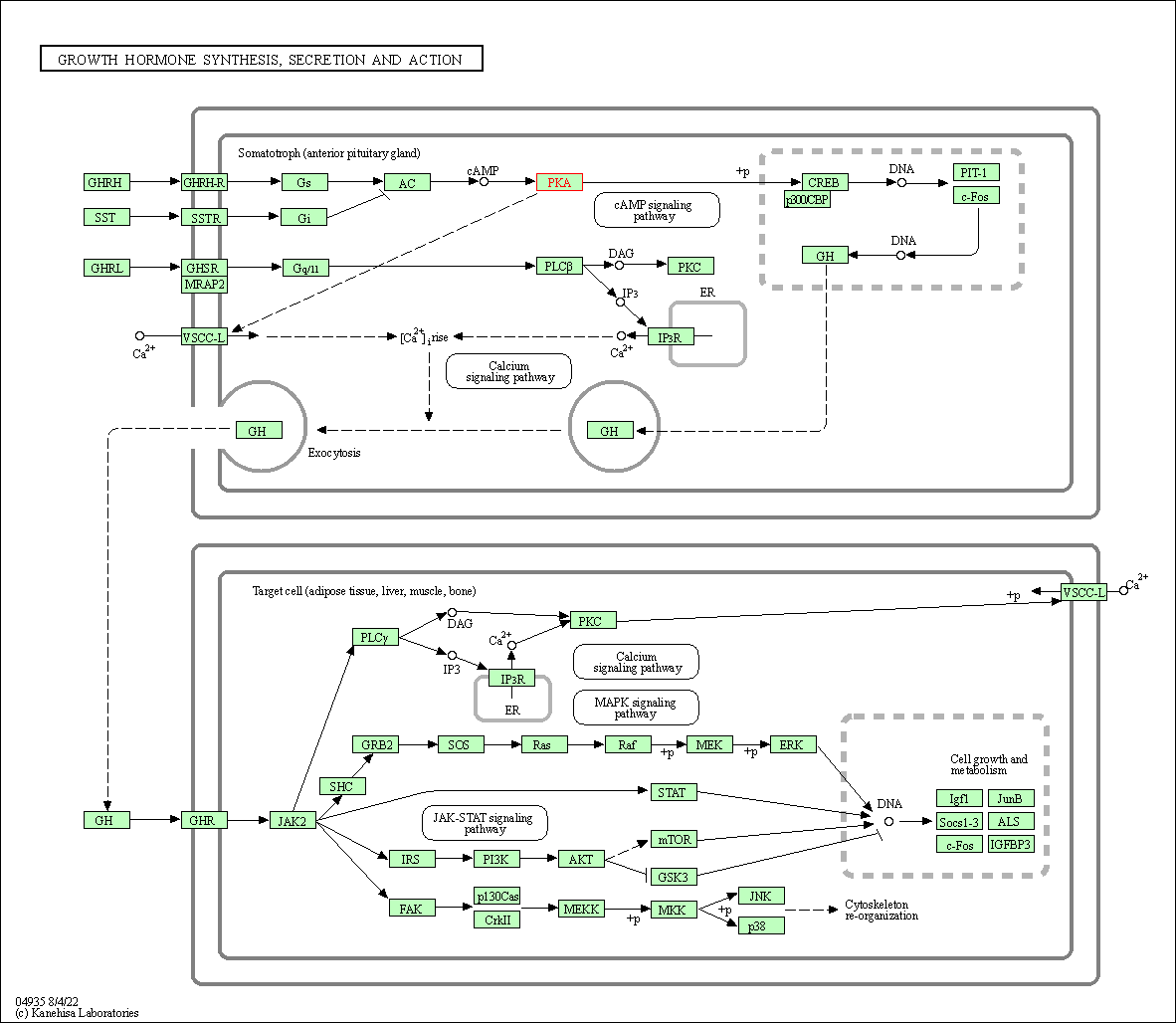
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Endocrine and other factor-regulated calcium reabsorption | hsa04961 | Affiliated Target |
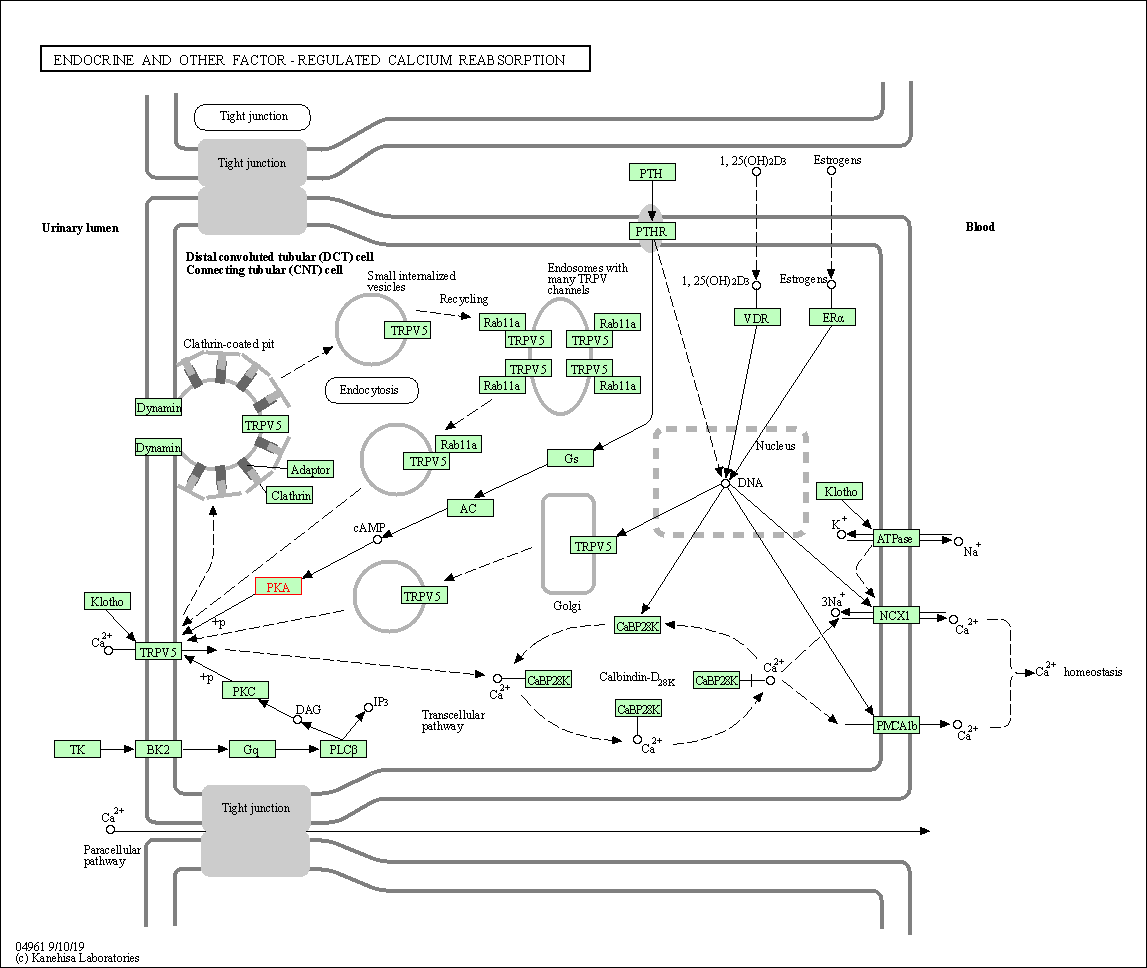
|
| Class: Organismal Systems => Excretory system | Pathway Hierarchy | ||
| Vasopressin-regulated water reabsorption | hsa04962 | Affiliated Target |
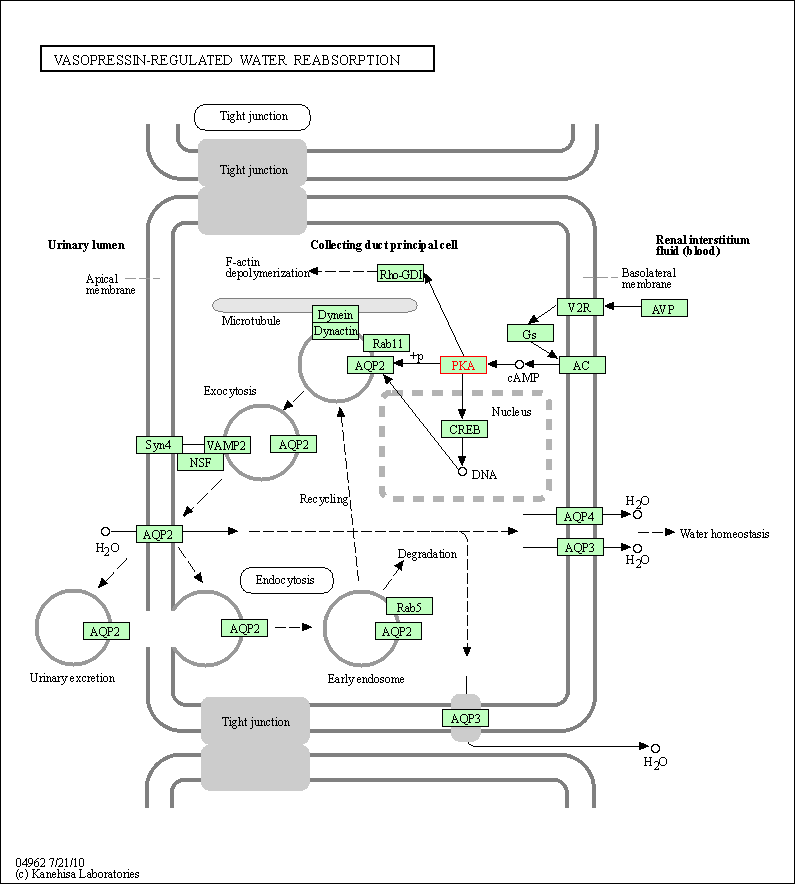
|
| Class: Organismal Systems => Excretory system | Pathway Hierarchy | ||
| Salivary secretion | hsa04970 | Affiliated Target |
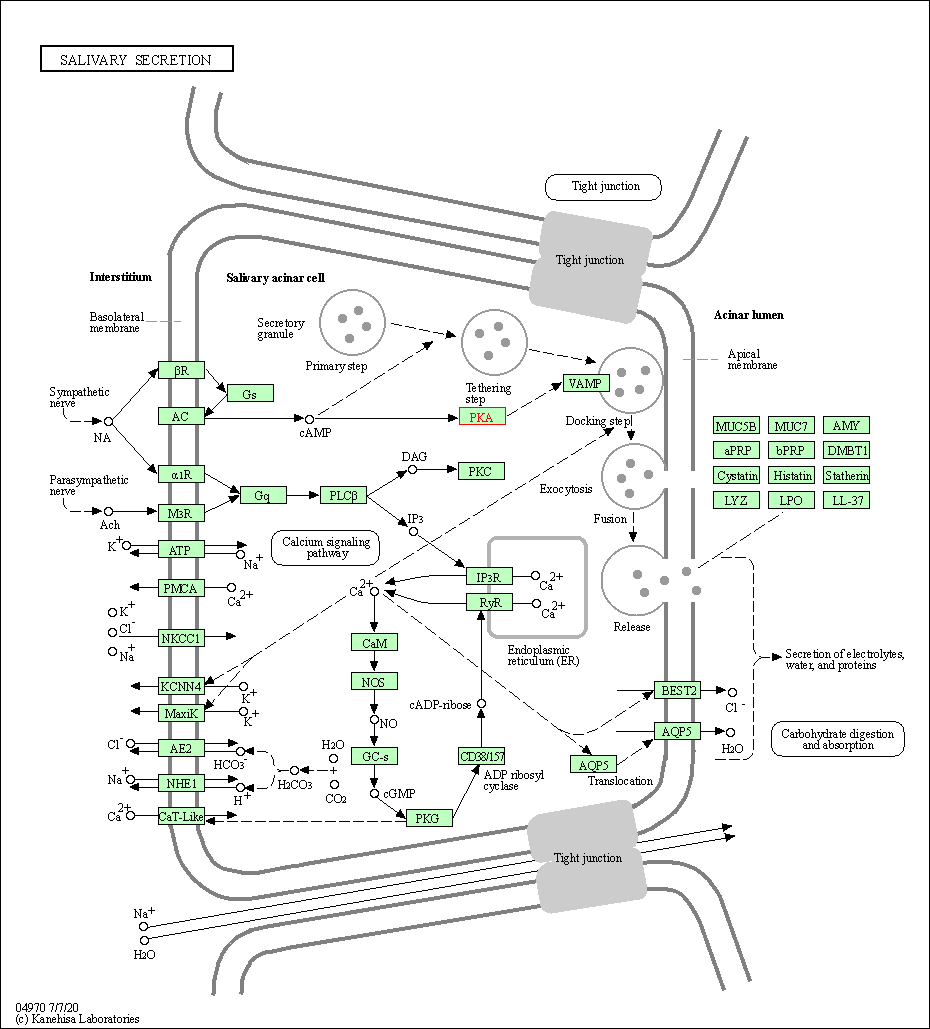
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Gastric acid secretion | hsa04971 | Affiliated Target |
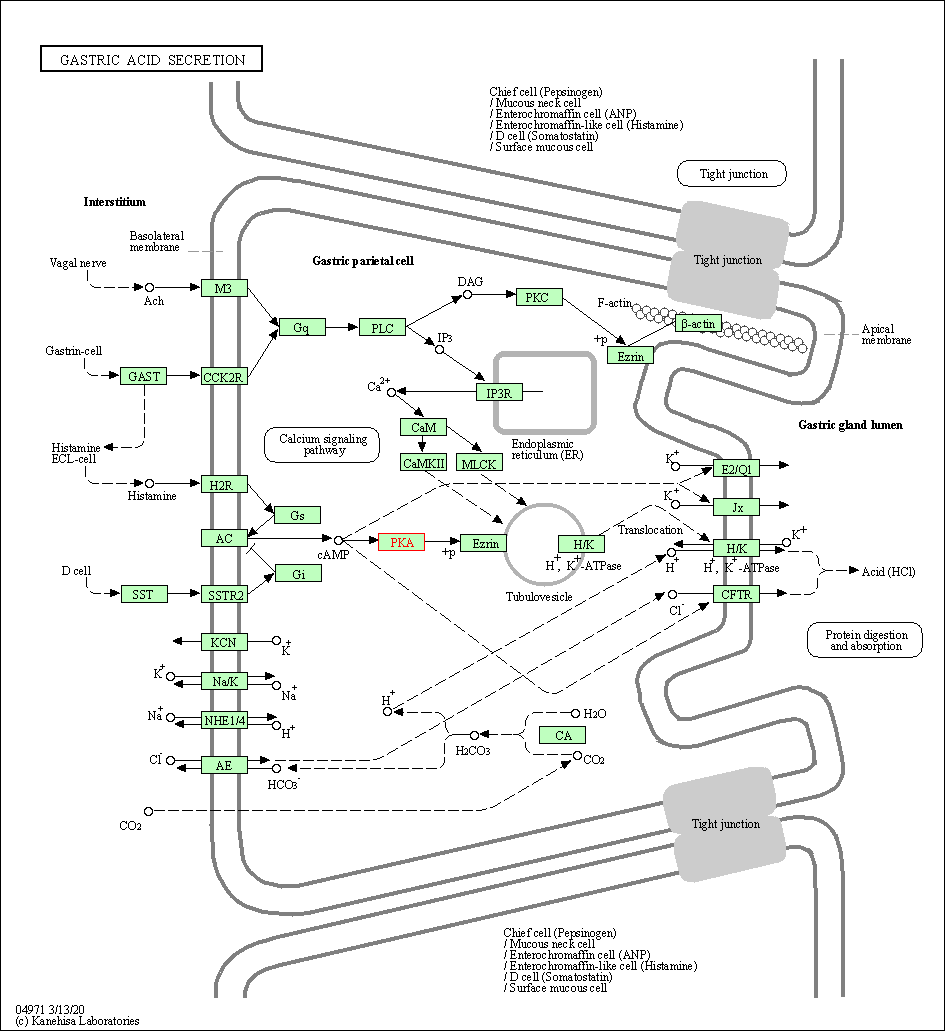
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Bile secretion | hsa04976 | Affiliated Target |
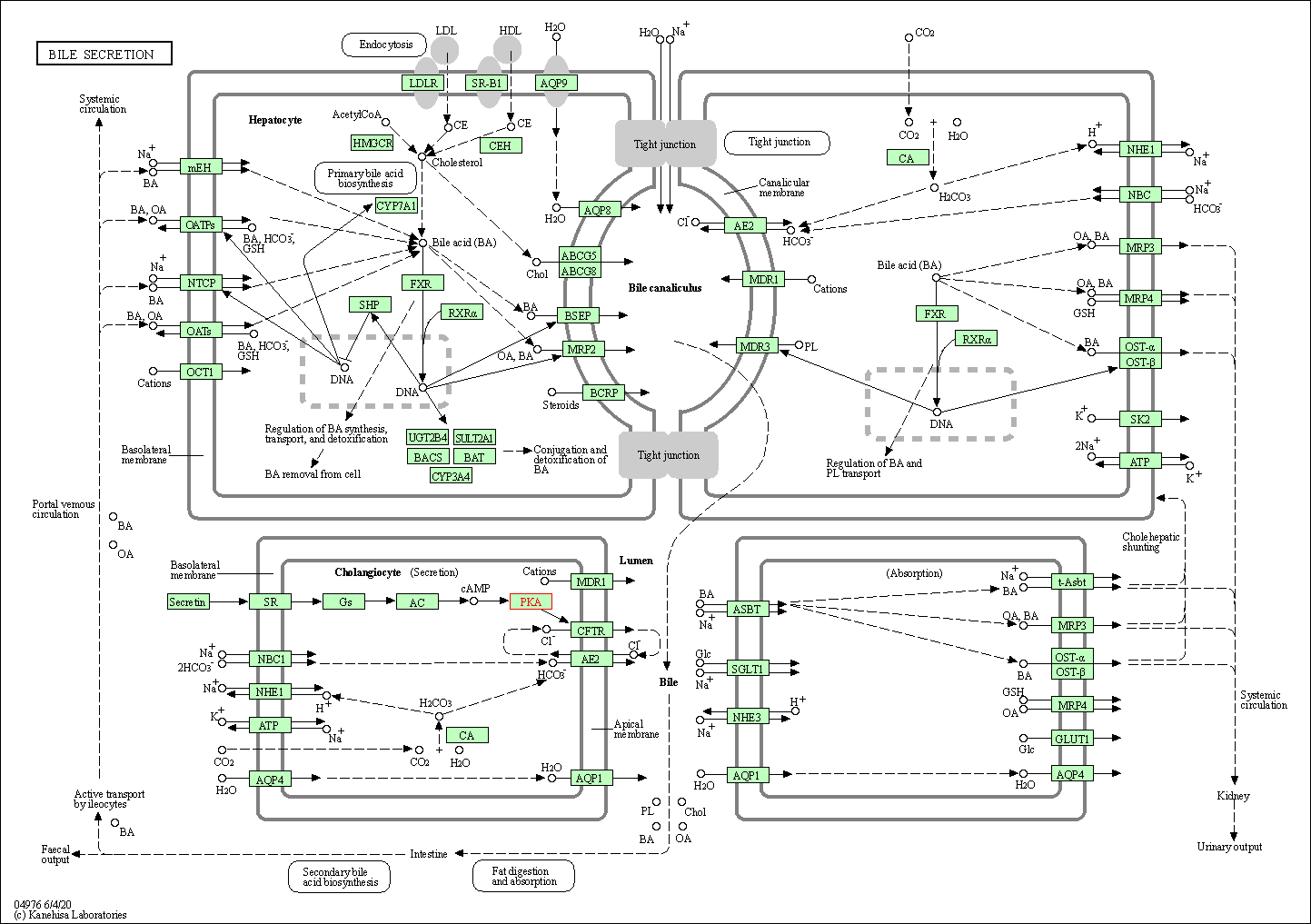
|
| Class: Organismal Systems => Digestive system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 20 | Degree centrality | 2.15E-03 | Betweenness centrality | 2.61E-03 |
|---|---|---|---|---|---|
| Closeness centrality | 2.41E-01 | Radiality | 1.42E+01 | Clustering coefficient | 1.11E-01 |
| Neighborhood connectivity | 2.42E+01 | Topological coefficient | 6.93E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | A SARS-CoV-2 Protein Interaction Map Reveals Targets for Drug Repurposing. Nature. 2020 Apr 30. doi: 10.1038/s41586-020-2286-9. | |||||
| REF 2 | Structural insights into mis-regulation of protein kinase A in human tumors. Proc Natl Acad Sci U S A. 2015 Feb 3;112(5):1374-9. | |||||
| REF 3 | Inhibitor induced structural effects involving Phe327 in AGC kinases | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

