| Drug General Information |
| Drug ID |
D0XF8W
|
| Drug Name |
Pyrazinamide |
|
| Synonyms |
Aldinamid; Aldinamide; Braccopiral; Corsazinmid; Dipimide; Eprazin; Farmizina; Isopas; Lynamide; Novamid; Pezetamid; Pharozinamide; Piraldina; Pirazimida; Pirazinamid; Pirazinamida; Pirazinamide; Prazina; Pyrafat; Pyramide; Pyrazide; Pyrazinamdie; Pyrazinamidum; Pyrazineamide; Pyrazinecarboxamide; Rozide; Tebrazid; Tebrazio; Tisamid; Unipyranamide; Zinamide; Zinastat; P ezetamid; Pirazinamide [DCIT]; Pyrazine carboxamide; Pyrazine carboxylamide; Pyrazinecarboxylic acid amide; Pyrazinoic acid am ide; Pyrazinoic acid amide; DRG 0124; MK 56; P 7136; Pyrazinamide BP 2000; T 165; AZT + Pyrazinamide combination; D-50; Pirazinamida [INN-Spanish]; Pms-Pyrazinamide; Pyrazinamide (TN); Pyrazinamidum [INN-Latin]; D-50 (VAN); Pyrazinamide [INN:BAN:JAN]; Pyrazine-2-carboxamide; Pyrazinamide (JP15/USP/INN); Pyrazinoic acid amide, Pezetamid, Pyrafat, Zinamide, Tebrazid, Pyrafat, Pyrazinamide; 2-Carbamylpyrazine; 2-carbamyl pyrazine |
| Drug Type |
Small molecular drug |
| Therapeutic Class |
Antitubercular Agents |
| Structure |
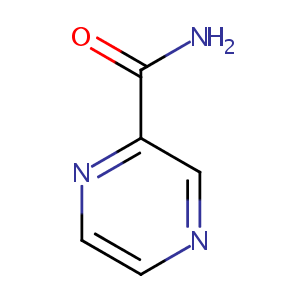
|
| Drug Resistance Mutations |
| Target Name |
Bacterial Pyrazinamidase (pncA) |
Target Info |
| Gene Name |
pncA |
| Uniprot ID |
PNCA_MYCTU |
| Species |
Mycobacterium tuberculosis |
| Reference Sequence |
MRALIIVDVQNDFCEGGSLAVTGGAALARAISDYLAEAADYHHVVATKDFHIDPGDHFSG
TPDYSSSWPPHCVSGTPGADFHPSLDTSAIEAVFYKGAYTGAYSGFEGVDENGTPLLNWL
RQRGVDEVDVVGIATDHCVRQTAEDAVRNGLATRVLVDLTAGVSADTTVAALEEMRTASV
ELVCSS [Mycobacterium tuberculosis]
|
| Targeted Disease |
Tuberculosis |
| Drug Resistance Mutations |
| Mutation info |
Frameshift: Plus 130 |
[1], [2], [3] |
|
| Mutation info |
Missense: I90S |
[4], [5], [6] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Frameshift: Plus 54 |
[2], [3], [7] |
|
| Mutation info |
Frameshift: Plus 96 |
[2], [7], [8] |
|
| Mutation info |
Missense: P69R |
[3], [6], [9] |
|
| Mutation info |
Missense: F58L |
[6], [8], [10] |
|
| Mutation info |
Missense: Q10P |
[6], [8], [11] |
|
| Mutation info |
Missense: A171P |
[12], [6] |
|
| Mutation info |
Frameshift: Plus 102 |
[1], [2] |
|
| Mutation info |
Frameshift: Plus 114 |
[1], [13] |
|
| Mutation info |
Frameshift: Plus 140 |
[1], [13] |
|
| Mutation info |
Missense: V155G |
[1], [6] |
|
| Mutation info |
Missense: A102T |
[14], [6] |
|
| Mutation info |
Missense: A102V |
[14], [6] |
|
| Mutation info |
Missense: A161P |
[14], [6] |
|
| Mutation info |
Missense: D49V |
[14], [6] |
|
| Mutation info |
Missense: D8Y |
[14], [6] |
|
| Mutation info |
Missense: I133T |
[14], [6] |
|
| Mutation info |
Missense: K96E |
[14], [6] |
|
| Mutation info |
Missense: L116R |
[14], [6] |
|
| Mutation info |
Missense: Q10K |
[14], [6] |
|
| Mutation info |
Missense: R154G |
[14], [6] |
|
| Mutation info |
Missense: S66P |
[14], [6] |
|
| Mutation info |
Missense: T160P |
[14], [6] |
|
| Mutation info |
Missense: T47S |
[14], [6] |
|
| Mutation info |
Missense: V125D |
[14], [6] |
|
| Mutation info |
Missense: A171T |
[15], [6] |
|
| Mutation info |
Missense: A171V |
[15], [6] |
|
| Mutation info |
Missense: D136H |
[15], [6] |
|
| Mutation info |
Missense: H51Y |
[15], [6] |
|
| Mutation info |
Missense: L172A |
[15], [6] |
|
| Mutation info |
Missense: L85R |
[15], [6] |
|
| Mutation info |
Missense: P62H |
[15], [6] |
|
| Mutation info |
Missense: Y103H |
[15], [6] |
|
| Mutation info |
Missense: A134V |
[10], [6] |
|
| Mutation info |
Missense: V7F |
[10], [6] |
|
| Mutation info |
Missense: A46E |
[13], [6] |
|
| Mutation info |
Missense: A46V |
[13], [6] |
|
| Mutation info |
Missense: C138S |
[13], [6] |
|
| Mutation info |
Missense: D53A |
[13], [6] |
|
| Mutation info |
Missense: G97S |
[13], [6] |
|
| Mutation info |
Missense: H71E |
[13], [6] |
|
| Mutation info |
Missense: H82R |
[13], [6] |
|
| Mutation info |
Frameshift: Plus 100 |
[13], [9] |
|
| Mutation info |
Frameshift: Plus 149 |
[13], [3] |
|
| Mutation info |
Missense: Q10R |
[13], [6] |
|
| Mutation info |
Missense: T168N |
[13], [6] |
|
| Mutation info |
Missense: V139M |
[13], [6] |
|
| Mutation info |
Missense: N118T |
[9], [6] |
|
| Mutation info |
Frameshift: Plus 123 |
[9], [2] |
|
| Mutation info |
Frameshift: Plus 134 |
[9], [16] |
|
| Mutation info |
Frameshift: Plus 1 |
[9], [3] |
|
| Mutation info |
Frameshift: Plus 24 |
[9], [16] |
|
| Mutation info |
Missense: T47P |
[9], [6] |
|
| Mutation info |
Missense: Y41H |
[9], [6] |
|
| Mutation info |
Missense: T76I |
[17], [6] |
|
| Mutation info |
Missense: G78D |
[11], [6] |
|
| Mutation info |
Missense: T135P |
[11], [6] |
|
| Mutation info |
Missense: V139L |
[11], [6] |
|
| Mutation info |
Nonsense: Y103 STOP |
[14], [11] |
|
| Mutation info |
Missense: A146T |
[2], [6] |
|
| Mutation info |
Missense: C14Y |
[2], [6] |
|
| Mutation info |
Missense: D136N |
[2], [6] |
|
| Mutation info |
Missense: D136Y |
[2], [6] |
|
| Mutation info |
Missense: D49A |
[2], [6] |
|
| Mutation info |
Missense: G23V |
[2], [6] |
|
| Mutation info |
Missense: K96Q |
[2], [6] |
|
| Mutation info |
Missense: L159R |
[2], [6] |
|
| Mutation info |
Missense: L4W |
[2], [6] |
|
| Mutation info |
Frameshift: Plus 127 |
[2], [18] |
| Mutation Frequency |
1 out of 48 patients |
|
| Mutation info |
Missense: V180F |
[2], [6] |
|
| Mutation info |
Missense: V45G |
[2], [6] |
|
| Mutation info |
Missense: V7D |
[2], [6] |
|
| Mutation info |
Missense: V9G |
[2], [6] |
|
| Mutation info |
Missense: W68G |
[2], [6] |
|
| Mutation info |
Missense: W68R |
[2], [6] |
|
| Mutation info |
Missense: W68S |
[2], [6] |
|
| Mutation info |
Missense: Y103S |
[2], [6] |
|
| Mutation info |
Missense: A3E |
[19], [6] |
|
| Mutation info |
Missense: C72W |
[19], [6] |
|
| Mutation info |
Missense: H51Q |
[19], [6] |
|
| Mutation info |
Missense: M175V |
[19], [6] |
|
| Mutation info |
Missense: R148S |
[17], [19] |
|
| Mutation info |
Missense: C138Y |
[7], [6] |
|
| Mutation info |
Missense: Q141P |
[7], [6] |
|
| Mutation info |
Missense: A26G |
[3], [6] |
|
| Mutation info |
Missense: C14R |
[3], [6] |
|
| Mutation info |
Missense: C72R |
[3], [6] |
|
| Mutation info |
Missense: D12A |
[3], [6] |
|
| Mutation info |
Missense: D12N |
[3], [6] |
|
| Mutation info |
Missense: G132S |
[3], [6] |
|
| Mutation info |
Missense: H137P |
[3], [6] |
|
| Mutation info |
Missense: I5S |
[3], [6] |
|
| Mutation info |
Missense: K96N |
[3], [6] |
|
| Mutation info |
Missense: L85P |
[3], [6] |
|
| Mutation info |
Missense: T142K |
[3], [6] |
|
| Mutation info |
Missense: T142M |
[3], [6] |
|
| Mutation info |
Frameshift: Plus 132 |
[16], [8] |
|
| Mutation info |
Missense: G17D |
[8], [6] |
|
| Mutation info |
Missense: L172P |
[8], [6] |
|
| Mutation info |
Missense: S67P |
[8], [6] |
|
| Mutation info |
Missense: V7G |
[8], [6] |
|
| Mutation info |
Missense: Y34S |
[8], [6] |
|
| Mutation info |
Nonsense: Y41 STOP |
[16], [8] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Nonsense: Y99 STOP |
[11], [8] |
|
| Mutation info |
Missense: F94P |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: H137R |
[16], [6] |
| Mutation Frequency |
2 out of 67 isolates |
|
| Mutation info |
Missense: H57D |
[16], [6] |
| Mutation Frequency |
2 out of 67 isolates |
|
| Mutation info |
Missense: L19P |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: P54L |
[5], [6] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: P54T |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: R121P |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: T47A |
[16], [6] |
| Mutation Frequency |
10 out of 67 isolates |
|
| Mutation info |
Missense: T76P |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: V139A |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: W119R |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: W68L |
[16], [6] |
| Mutation Frequency |
1 out of 67 isolates |
|
| Mutation info |
Missense: F80V |
[12] |
|
| Mutation info |
Missense: A3P |
[1] |
|
| Mutation info |
Missense: F13S |
[1] |
|
| Mutation info |
Missense: L182S |
[1] |
|
| Mutation info |
Missense: P69L |
[1] |
|
| Mutation info |
Frameshift: Plus 145 |
[1] |
|
| Mutation info |
Frameshift: Plus 64 |
[1] |
|
| Mutation info |
Frameshift: Plus 72 |
[1] |
|
| Mutation info |
Missense: T61P |
[1] |
|
| Mutation info |
Missense: T87M |
[1] |
|
| Mutation info |
Missense: Y103C |
[1] |
|
| Mutation info |
Missense: D49G |
[14] |
|
| Mutation info |
Frameshift: Plus 126 |
[14] |
|
| Mutation info |
Frameshift: Plus 28 |
[14] |
|
| Mutation info |
Nonsense: Q10 STOP |
[14] |
|
| Mutation info |
Missense: L120P |
[20] |
|
| Mutation info |
Missense: M1T |
[20] |
|
| Mutation info |
Frameshift: Plus 136 |
[15] |
|
| Mutation info |
Frameshift: Plus 171 |
[15] |
|
| Mutation info |
Frameshift: Plus 65 |
[15] |
|
| Mutation info |
Frameshift: Plus 74 |
[15] |
|
| Mutation info |
Missense: T142P |
[10] |
|
| Mutation info |
Missense: V155A |
[10] |
|
| Mutation info |
Missense: V7I |
[10] |
|
| Mutation info |
Frameshift: Plus 143 |
[13] |
|
| Mutation info |
Frameshift: Plus 17 |
[13] |
|
| Mutation info |
Missense: R140S |
[13] |
|
| Mutation info |
Nonsense: W68 STOP |
[13] |
|
| Mutation info |
Frameshift: Plus 167 |
[9] |
|
| Mutation info |
Frameshift: Plus 20 |
[9] |
|
| Mutation info |
Nonsense: S88 STOP |
[9] |
|
| Mutation info |
Missense: H57P |
[17] |
|
| Mutation info |
Missense: H82D |
[17] |
|
| Mutation info |
Frameshift: Plus 80 |
[17] |
|
| Mutation info |
Missense: S59P |
[17] |
|
| Mutation info |
Insertion: E/V/D129 |
[11] |
|
| Mutation info |
Missense: G97D |
[11] |
|
| Mutation info |
Missense: H71R |
[11] |
|
| Mutation info |
Missense: I133N |
[11] |
|
| Mutation info |
Missense: L4S |
[11] |
|
| Mutation info |
Missense: P62R |
[11] |
|
| Mutation info |
Frameshift: Plus 35 |
[11] |
|
| Mutation info |
Missense: T153N |
[11] |
|
| Mutation info |
Missense: V139G |
[11] |
|
| Mutation info |
Frameshift: Plus 29 |
[4] |
|
| Mutation info |
Missense: A25E |
[2] |
|
| Mutation info |
Missense: A28D |
[2] |
|
| Mutation info |
Missense: D8G |
[2] |
|
| Mutation info |
Missense: M1I |
[2] |
|
| Mutation info |
Frameshift: Plus 129 |
[2] |
|
| Mutation info |
Frameshift: Plus 131 |
[2] |
|
| Mutation info |
Frameshift: Plus 135 |
[2] |
|
| Mutation info |
Frameshift: Plus 138 |
[2] |
|
| Mutation info |
Frameshift: Plus 151 |
[2] |
|
| Mutation info |
Frameshift: Plus 155 |
[2] |
|
| Mutation info |
Frameshift: Plus 160 |
[2] |
|
| Mutation info |
Frameshift: Plus 177 |
[2] |
|
| Mutation info |
Frameshift: Plus 50 |
[2] |
|
| Mutation info |
Frameshift: Plus 53 |
[2] |
|
| Mutation info |
Nonsense: Q141 STOP |
[2] |
|
| Mutation info |
Missense: V9A |
[2] |
|
| Mutation info |
Nonsense: W119 STOP |
[2] |
|
| Mutation info |
Missense: Y64D |
[2] |
|
| Mutation info |
Missense: A171E |
[21] |
|
| Mutation info |
Missense: H51N |
[21] |
|
| Mutation info |
Missense: H82L |
[21] |
|
| Mutation info |
Missense: V128G |
[21] |
|
| Mutation info |
Missense: A46P |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: C14H |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: C72Y |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: D136G |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: D49H |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: E181D |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Nonsense: E37 STOP |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: G107K |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: H57Y |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: H71D |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: H71Y |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: L116V |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: L159P |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: L35R |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Frameshift: Plus 58 |
[5] |
| Mutation Frequency |
63 out of 68 patients in all pncA mutations |
|
| Mutation info |
Missense: S179R |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: S185T |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: V9L |
[5] |
| Mutation Frequency |
63 out of 68 isolates in all pncA mutations |
|
| Mutation info |
Missense: A79G |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: G162D |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: G17V |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: H57L |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: L19R |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: M75T |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: P62L |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Frameshift: Plus 16 |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Frameshift: Plus 22 |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: S185I |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: V21G |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: V9S |
[22] |
| Mutation Frequency |
25% of the patients in all pncA mutations |
|
| Mutation info |
Missense: D53N |
[19] |
|
| Mutation info |
Frameshift: Plus 73 |
[19] |
|
| Mutation info |
Frameshift: Plus 147 |
[18] |
| Mutation Frequency |
3 out of 48 patients |
|
| Mutation info |
Missense: V125G |
[18] |
| Mutation Frequency |
19 out of 48 isolates |
|
| Mutation info |
Nonsense: Y95 STOP |
[18] |
|
| Mutation info |
Missense: L35P |
[23] |
|
| Mutation info |
Missense: W119C |
[23] |
|
| Mutation info |
Missense: D63H |
[7] |
|
| Mutation info |
Missense: D8E |
[3] |
|
| Mutation info |
Missense: K96T |
[3] |
|
| Mutation info |
Frameshift: Plus 104 |
[3] |
|
| Mutation info |
Frameshift: Plus 10 |
[3] |
|
| Mutation info |
Frameshift: Plus 139 |
[3] |
|
| Mutation info |
Frameshift: Plus 148 |
[3] |
|
| Mutation info |
Frameshift: Plus 158 |
[3] |
|
| Mutation info |
Frameshift: Plus 173 |
[3] |
|
| Mutation info |
Frameshift: Plus 33 |
[3] |
|
| Mutation info |
Frameshift: Plus 34 |
[3] |
|
| Mutation info |
Insertion: R/S/M129 |
[16] |
|
| Mutation info |
Missense: A146V |
[8] |
|
| Mutation info |
Missense: D63G |
[8] |
|
| Mutation info |
Missense: G132D |
[8] |
|
| Mutation info |
Missense: H43P |
[8] |
|
| Mutation info |
Missense: H51P |
[8] |
|
| Mutation info |
Missense: H51R |
[8] |
|
| Mutation info |
Frameshift: Plus 46 |
[8] |
|
| Mutation info |
Missense: S104R |
[8] |
|
| Mutation info |
Missense: T114P |
[8] |
|
| Mutation info |
Missense: T142A |
[8] |
|
| Mutation info |
Missense: V130G |
[8] |
|
| Mutation info |
Missense: D12E |
[6] |
|
| Mutation info |
Missense: L27P |
[6] |
|
| Mutation info |
Missense: M1S |
[6] |
|
| Mutation info |
Missense: S104C |
[6] |
|
| References |
| REF 1 |
Characterization of new mutations in pyrazinamide-resistant strains of Mycobacterium tuberculosis and identification of conserved regions important for the catalytic activity of the pyrazinamidase PncA. Antimicrob Agents Chemother. 1999 Jul;43(7):1761-3.
|
| REF 2 |
Characterization of pncA mutations of pyrazinamide-resistant Mycobacterium tuberculosis in Korea. J Korean Med Sci. 2001 Oct;16(5):537-43.
|
| REF 3 |
Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1997 Mar;41(3):540-3.
|
| REF 4 |
Pyrazinamide-monoresistant Mycobacterium tuberculosis in the United States. J Clin Microbiol. 2001 Feb;39(2):647-50.
|
| REF 5 |
Frequency and implications of pyrazinamide resistance in managing previously treated tuberculosis patients. Int J Tuberc Lung Dis. 2006 Jul;10(7):802-7.
|
| REF 6 |
The comprehensive antibiotic resistance database. Antimicrob Agents Chemother. 2013 Jul;57(7):3348-57.
|
| REF 7 |
Mutations in pncA, a gene encoding pyrazinamidase/nicotinamidase, cause resistance to the antituberculous drug pyrazinamide in tubercle bacillus. Nat Med. 1996 Jun;2(6):662-7.
|
| REF 8 |
Mutation in pncA is a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis. Tuber Lung Dis. 1997;78(2):117-22.
|
| REF 9 |
Molecular characterization of pncA gene mutations in Mycobacterium tuberculosis clinical isolates from China. Epidemiol Infect. 2000 Apr;124(2):227-32.
|
| REF 10 |
Genotypic characterization of drug-resistant Mycobacterium tuberculosis isolates from Peru. Tuber Lung Dis. 1998;79(2):111-8.
|
| REF 11 |
Characterization of spontaneous, In vitro-selected, rifampin-resistant mutants of Mycobacterium tuberculosis strain H37Rv. Antimicrob Agents Chemother. 2000 Dec;44(12):3298-301.
|
| REF 12 |
Novel pncA Mutations in Pyrazinamide-Resistant Isolates of Mycobacterium tuberculosis. Mol Diagn. 1998 Dec;3(4):229-231.
|
| REF 13 |
pncA mutations as a major mechanism of pyrazinamide resistance in Mycobacterium tuberculosis: spread of a monoresistant strain in Quebec, Canada. Antimicrob Agents Chemother. 2000 Mar;44(3):528-32.
|
| REF 14 |
pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis isolates from northwestern Russia. Antimicrob Agents Chemother. 1999 Jul;43(7):1764-6.
|
| REF 15 |
Relationship between pyrazinamide resistance, loss of pyrazinamidase activity, and mutations in the pncA locus in multidrug-resistant clinical isolates of Mycobacterium tuberculosis. Antimicrob Agents Chemother. 1999 Sep;43(9):2317-9.
|
| REF 16 |
Mutations associated with pyrazinamide resistance in pncA of Mycobacterium tuberculosis complex organisms. Antimicrob Agents Chemother. 1997 Mar;41(3):636-40.
|
| REF 17 |
Simultaneous identification and typing of multi-drug-resistant Mycobacterium tuberculosis isolates by analysis of pncA and rpoB. J Med Microbiol. 2000 Jul;49(7):651-6.
|
| REF 18 |
Multidrug-resistant tuberculosis in Lisbon, Portugal: a molecular epidemiological perspective. Microb Drug Resist. 2008 Jun;14(2):133-43.
|
| REF 19 |
Development and evaluation of a line probe assay for rapid identification of pncA mutations in pyrazinamide-resistant mycobacterium tuberculosis st... J Clin Microbiol. 2007 Sep;45(9):2802-7.
|
| REF 20 |
Discrimination of multidrug-resistant Mycobacterium tuberculosis IS6110 fingerprint subclusters by rpoB gene mutation analysis. J Clin Microbiol. 1999 Sep;37(9):3022-4.
|
| REF 21 |
Characterization of pncA mutations in pyrazinamide-resistant Mycobacterium tuberculosis in Brazil. Antimicrob Agents Chemother. 2005 Jan;49(1):444-6.
|
| REF 22 |
Genetic and phenotypic characterization of drug-resistant Mycobacterium tuberculosis isolates in Hong Kong. J Antimicrob Chemother. 2007 May;59(5):866-73.
|
| REF 23 |
In-depth molecular characterization of Mycobacterium tuberculosis from New Delhi--predominance of drug resistant isolates of the 'modern' (TbD1) type. PLoS One. 2009;4(2):e4540.
|


