Target Information
| Target General Information | Top | |||||
|---|---|---|---|---|---|---|
| Target ID |
T99685
|
|||||
| Target Name |
HUMAN mammalian target of rapamycin (mTOR)
|
|||||
| Synonyms |
Target of rapamycin; TOR kinase; Rapamycin target protein 1; Rapamycin target protein; Rapamycin and FKBP12 target 1; RAPT1; RAFT1; Mechanistic target of rapamycin; Mammalian target of rapamycin; FRAP2; FRAP1; FRAP; FKBP12-rapamycin complex-associated protein; FKBP-rapamycin associated protein; FK506-binding protein 12-rapamycin complex-associated protein 1
Click to Show/Hide
|
|||||
| Gene Name |
MTOR
|
|||||
| Disease | [+] 2 Target-related Diseases | + | ||||
| 1 | Advanced/metastatic breast cancer [ICD-11: 2C60] | |||||
| 2 | Lymphangioleiomyomatosis [ICD-11: CB07] | |||||
| Function |
MTOR directly or indirectly regulates the phosphorylation of at least 800 proteins. Functions as part of 2 structurally and functionally distinct signaling complexes mTORC1 and mTORC2 (mTOR complex 1 and 2). Activated mTORC1 up-regulates protein synthesis by phosphorylating key regulators of mRNA translation and ribosome synthesis. This includes phosphorylation of EIF4EBP1 and release of its inhibition toward the elongation initiation factor 4E (eiF4E). Moreover, phosphorylates and activates RPS6KB1 and RPS6KB2 that promote protein synthesis by modulating the activity of their downstream targets including ribosomal protein S6, eukaryotic translation initiation factor EIF4B, and the inhibitor of translation initiation PDCD4. Stimulates the pyrimidine biosynthesis pathway, both by acute regulation through RPS6KB1-mediated phosphorylation of the biosynthetic enzyme CAD, and delayed regulation, through transcriptional enhancement of the pentose phosphate pathway which produces 5-phosphoribosyl-1-pyrophosphate (PRPP), an allosteric activator of CAD at a later step in synthesis, this function is dependent on the mTORC1 complex. Regulates ribosome synthesis by activating RNA polymerase III-dependent transcription through phosphorylation and inhibition of MAF1 an RNA polymerase III-repressor. In parallel to protein synthesis, also regulates lipid synthesis through SREBF1/SREBP1 and LPIN1. To maintain energy homeostasis mTORC1 may also regulate mitochondrial biogenesis through regulation of PPARGC1A. mTORC1 also negatively regulates autophagy through phosphorylation of ULK1. Under nutrient sufficiency, phosphorylates ULK1 at 'Ser-758', disrupting the interaction with AMPK and preventing activation of ULK1. Also prevents autophagy through phosphorylation of the autophagy inhibitor DAP. Also prevents autophagy by phosphorylating RUBCNL/Pacer under nutrient-rich conditions. mTORC1 exerts a feedback control on upstream growth factor signaling that includes phosphorylation and activation of GRB10 a INSR-dependent signaling suppressor. Among other potential targets mTORC1 may phosphorylate CLIP1 and regulate microtubules. As part of the mTORC2 complex MTOR may regulate other cellular processes including survival and organization of the cytoskeleton. Plays a critical role in the phosphorylation at 'Ser-473' of AKT1, a pro-survival effector of phosphoinositide 3-kinase, facilitating its activation by PDK1. mTORC2 may regulate the actin cytoskeleton, through phosphorylation of PRKCA, PXN and activation of the Rho-type guanine nucleotide exchange factors RHOA and RAC1A or RAC1B. mTORC2 also regulates the phosphorylation of SGK1 at 'Ser-422'. Regulates osteoclastogenesis by adjusting the expression of CEBPB isoforms. Plays an important regulatory role in the circadian clock function; regulates period length and rhythm amplitude of the suprachiasmatic nucleus (SCN) and liver clocks. Serine/threonine protein kinase which is a central regulator of cellular metabolism, growth and survival in response to hormones, growth factors, nutrients, energy and stress signals.
Click to Show/Hide
|
|||||
| BioChemical Class |
Kinase
|
|||||
| UniProt ID | ||||||
| EC Number |
EC 2.7.11.1
|
|||||
| Sequence |
MLGTGPAAATTAATTSSNVSVLQQFASGLKSRNEETRAKAAKELQHYVTMELREMSQEES
TRFYDQLNHHIFELVSSSDANERKGGILAIASLIGVEGGNATRIGRFANYLRNLLPSNDP VVMEMASKAIGRLAMAGDTFTAEYVEFEVKRALEWLGADRNEGRRHAAVLVLRELAISVP TFFFQQVQPFFDNIFVAVWDPKQAIREGAVAALRACLILTTQREPKEMQKPQWYRHTFEE AEKGFDETLAKEKGMNRDDRIHGALLILNELVRISSMEGERLREEMEEITQQQLVHDKYC KDLMGFGTKPRHITPFTSFQAVQPQQSNALVGLLGYSSHQGLMGFGTSPSPAKSTLVESR CCRDLMEEKFDQVCQWVLKCRNSKNSLIQMTILNLLPRLAAFRPSAFTDTQYLQDTMNHV LSCVKKEKERTAAFQALGLLSVAVRSEFKVYLPRVLDIIRAALPPKDFAHKRQKAMQVDA TVFTCISMLARAMGPGIQQDIKELLEPMLAVGLSPALTAVLYDLSRQIPQLKKDIQDGLL KMLSLVLMHKPLRHPGMPKGLAHQLASPGLTTLPEASDVGSITLALRTLGSFEFEGHSLT QFVRHCADHFLNSEHKEIRMEAARTCSRLLTPSIHLISGHAHVVSQTAVQVVADVLSKLL VVGITDPDPDIRYCVLASLDERFDAHLAQAENLQALFVALNDQVFEIRELAICTVGRLSS MNPAFVMPFLRKMLIQILTELEHSGIGRIKEQSARMLGHLVSNAPRLIRPYMEPILKALI LKLKDPDPDPNPGVINNVLATIGELAQVSGLEMRKWVDELFIIIMDMLQDSSLLAKRQVA LWTLGQLVASTGYVVEPYRKYPTLLEVLLNFLKTEQNQGTRREAIRVLGLLGALDPYKHK VNIGMIDQSRDASAVSLSESKSSQDSSDYSTSEMLVNMGNLPLDEFYPAVSMVALMRIFR DQSLSHHHTMVVQAITFIFKSLGLKCVQFLPQVMPTFLNVIRVCDGAIREFLFQQLGMLV SFVKSHIRPYMDEIVTLMREFWVMNTSIQSTIILLIEQIVVALGGEFKLYLPQLIPHMLR VFMHDNSPGRIVSIKLLAAIQLFGANLDDYLHLLLPPIVKLFDAPEAPLPSRKAALETVD RLTESLDFTDYASRIIHPIVRTLDQSPELRSTAMDTLSSLVFQLGKKYQIFIPMVNKVLV RHRINHQRYDVLICRIVKGYTLADEEEDPLIYQHRMLRSGQGDALASGPVETGPMKKLHV STINLQKAWGAARRVSKDDWLEWLRRLSLELLKDSSSPSLRSCWALAQAYNPMARDLFNA AFVSCWSELNEDQQDELIRSIELALTSQDIAEVTQTLLNLAEFMEHSDKGPLPLRDDNGI VLLGERAAKCRAYAKALHYKELEFQKGPTPAILESLISINNKLQQPEAAAGVLEYAMKHF GELEIQATWYEKLHEWEDALVAYDKKMDTNKDDPELMLGRMRCLEALGEWGQLHQQCCEK WTLVNDETQAKMARMAAAAAWGLGQWDSMEEYTCMIPRDTHDGAFYRAVLALHQDLFSLA QQCIDKARDLLDAELTAMAGESYSRAYGAMVSCHMLSELEEVIQYKLVPERREIIRQIWW ERLQGCQRIVEDWQKILMVRSLVVSPHEDMRTWLKYASLCGKSGRLALAHKTLVLLLGVD PSRQLDHPLPTVHPQVTYAYMKNMWKSARKIDAFQHMQHFVQTMQQQAQHAIATEDQQHK QELHKLMARCFLKLGEWQLNLQGINESTIPKVLQYYSAATEHDRSWYKAWHAWAVMNFEA VLHYKHQNQARDEKKKLRHASGANITNATTAATTAATATTTASTEGSNSESEAESTENSP TPSPLQKKVTEDLSKTLLMYTVPAVQGFFRSISLSRGNNLQDTLRVLTLWFDYGHWPDVN EALVEGVKAIQIDTWLQVIPQLIARIDTPRPLVGRLIHQLLTDIGRYHPQALIYPLTVAS KSTTTARHNAANKILKNMCEHSNTLVQQAMMVSEELIRVAILWHEMWHEGLEEASRLYFG ERNVKGMFEVLEPLHAMMERGPQTLKETSFNQAYGRDLMEAQEWCRKYMKSGNVKDLTQA WDLYYHVFRRISKQLPQLTSLELQYVSPKLLMCRDLELAVPGTYDPNQPIIRIQSIAPSL QVITSKQRPRKLTLMGSNGHEFVFLLKGHEDLRQDERVMQLFGLVNTLLANDPTSLRKNL SIQRYAVIPLSTNSGLIGWVPHCDTLHALIRDYREKKKILLNIEHRIMLRMAPDYDHLTL MQKVEVFEHAVNNTAGDDLAKLLWLKSPSSEVWFDRRTNYTRSLAVMSMVGYILGLGDRH PSNLMLDRLSGKILHIDFGDCFEVAMTREKFPEKIPFRLTRMLTNAMEVTGLDGNYRITC HTVMEVLREHKDSVMAVLEAFVYDPLLNWRLMDTNTKGNKRSRTRTDSYSAGQSVEILDG VELGEPAHKKTGTTVPESIHSFIGDGLVKPEALNKKAIQIINRVRDKLTGRDFSHDDTLD VPTQVELLIKQATSHENLCQCYIGWCPFW Click to Show/Hide
|
|||||
| 3D Structure | Click to Show 3D Structure of This Target | PDB | ||||
| Drugs and Modes of Action | Top | |||||
|---|---|---|---|---|---|---|
| Investigative Agents | [+] 3 | + | ||||
| 1 | Everolimus | Drug Info | Approved | Advanced/metastatic breast cancer | [2] | |
| 2 | Rapamycin | Drug Info | Approved | Lymphangioleiomyomatosis | [2] | |
| 3 | Sapanisertib | Drug Info | Phase 2 | Breast cancer | [4] | |
| Drugs in Phase 2 Trial | [+] 1 | + | ||||
| 1 | Sirolimus | Drug Info | Approved | Lymphangioleiomyomatosis | [3] | |
| Mode of Action | [+] 1 Modes of Action | + | ||||
| Inhibitor | [+] 4 Inhibitor drugs | + | ||||
| 1 | Sirolimus | Drug Info | [5] | |||
| 2 | Everolimus | Drug Info | [1] | |||
| 3 | Rapamycin | Drug Info | [1] | |||
| 4 | Sapanisertib | Drug Info | [6] | |||
| Cell-based Target Expression Variations | Top | |||||
|---|---|---|---|---|---|---|
| Cell-based Target Expression Variations | ||||||
| Drug Binding Sites of Target | Top | |||||
|---|---|---|---|---|---|---|
| Ligand Name: Sirolimus | Ligand Info | |||||
| Structure Description | Co-crystal structure of the PPIase domain of FKBP51, Rapamycin and the FRB fragment of mTOR | PDB:4DRI | ||||
| Method | X-ray diffraction | Resolution | 1.45 Å | Mutation | No | [7] |
| PDB Sequence |
GAMDPEFMEM
2026 WHEGLEEASR2036 LYFGERNVKG2046 MFEVLEPLHA2056 MMERGPQTLK2066 ETSFNQAYGR 2076 DLMEAQEWCR2086 KYMKSGNVKD2096 LTQAWDLYYH2106 VFRRIS
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Ligand Name: Myo-inositol hexaphosphate | Ligand Info | |||||
| Structure Description | cryo-EM structure of human mTOR complex 2, focused on one half | PDB:6ZWO | ||||
| Method | Electron microscopy | Resolution | 3.00 Å | Mutation | No | [8] |
| PDB Sequence |
NPGVINNVLA
800 TIGELAQVSG810 LEMRKWVDEL820 FIIIMDMLQD830 SSLLAKRQVA840 LWTLGQLVAS 850 TGYVVEPYRK860 YPTLLEVLLN870 FLKTEQNQGT880 RREAIRVLGL890 LGALDPYKHK 900 VNISDYSTSE933 MLVNMGNLPL943 DEFYPAVSMV953 ALMRIFRDQS963 LSHHHTMVVQ 973 AITFIFKSLG983 LKCVQFLPQV993 MPTFLNVIRV1003 CDGAIREFLF1013 QQLGMLVSFV 1023 KSHIRPYMDE1033 IVTLMREFWV1043 MNTSIQSTII1053 LLIEQIVVAL1063 GGEFKLYLPQ 1073 LIPHMLRVFM1083 HDNSPGRIVS1093 IKLLAAIQLF1103 GANLDDYLHL1113 LLPPIVKLFD 1123 APEAPLPSRK1133 AALETVDRLT1143 ESLDFTDYAS1153 RIIHPIVRTL1163 DQSPELRSTA 1173 MDTLSSLVFQ1183 LGKKYQIFIP1193 MVNKVLVRHR1203 INHQRYDVLI1213 CRIVKGYTLA 1223 DEEEDPLIYQ1233 HRMLRINLQK1267 AWGAARRVSK1277 DDWLEWLRRL1287 SLELLKDSSS 1297 PSLRSCWALA1307 QAYNPMARDL1317 FNAAFVSCWS1327 ELNEDQQDEL1337 IRSIELALTS 1347 QDIAEVTQTL1357 LNLAEFMEHS1367 DKGPLPLRDD1377 NGIVLLGERA1387 AKCRAYAKAL 1397 HYKELEFQKG1407 PTPAILESLI1417 SINNKLQQPE1427 AAAGVLEYAM1437 KHFGELEIQA 1447 TWYEKLHEWE1457 DALVAYDKKM1467 DTNKDDPELM1477 LGRMRCLEAL1487 GEWGQLHQQC 1497 CEKWTLVNDE1507 TQAKMARMAA1517 AAAWGLGQWD1527 SMEEYTCMIP1537 RDTHDGAFYR 1547 AVLALHQDLF1557 SLAQQCIDKA1567 RDLLDAELTA1577 MAGESYSRAY1587 GAMVSCHMLS 1597 ELEEVIQYKL1607 VPERREIIRQ1617 IWWERLQGCQ1627 RIVEDWQKIL1637 MVRSLVVSPH 1647 EDMRTWLKYA1657 SLCGKSGRLA1667 LAHKTLVLLL1677 GVDPSRQLDH1687 PLPTVHPQVT 1697 YAYMKNMWKS1707 ARKIDAFQHM1717 QHFVQTMQQQ1727 AQHAIATEDQ1737 QHKQELHKLM 1747 ARCFLKLGEW1757 QLNLQGINES1767 TIPKVLQYYS1777 AATEHDRSWY1787 KAWHAWAVMN 1797 FEAVLHYKHQ1807 NQALSKTLLM1879 YTVPAVQGFF1889 RSISLSRGNN1899 LQDTLRVLTL 1909 WFDYGHWPDV1919 NEALVEGVKA1929 IQIDTWLQVI1939 PQLIARIDTP1949 RPLVGRLIHQ 1959 LLTDIGRYHP1969 QALIYPLTVA1979 SKSTTTARHN1989 AANKILKNMC1999 EHSNTLVQQA 2009 MMVSEELIRV2019 AILWHEMWHE2029 GLEEASRLYF2039 GERNVKGMFE2049 VLEPLHAMME 2059 RGPQTLKETS2069 FNQAYGRDLM2079 EAQEWCRKYM2089 KSGNVKDLTQ2099 AWDLYYHVFR 2109 RISKQLPQLT2119 SLELQYVSPK2129 LLMCRDLELA2139 VPGTYDPNQP2149 IIRIQSIAPS 2159 LQVITSKQRP2169 RKLTLMGSNG2179 HEFVFLLKGH2189 EDLRQDERVM2199 QLFGLVNTLL 2209 ANDPTSLRKN2219 LSIQRYAVIP2229 LSTNSGLIGW2239 VPHCDTLHAL2249 IRDYREKKKI 2259 LLNIEHRIML2269 RMAPDYDHLT2279 LMQKVEVFEH2289 AVNNTAGDDL2299 AKLLWLKSPS 2309 SEVWFDRRTN2319 YTRSLAVMSM2329 VGYILGLGDR2339 HPSNLMLDRL2349 SGKILHIDFG 2359 DCFEVAMTRE2369 KFPEKIPFRL2379 TRMLTNAMEV2389 TGLDGNYRIT2399 CHTVMEVLRE 2409 HKDSVMAVLE2419 AFVYDPLLNW2429 RLMDALNKKA2497 IQIINRVRDK2507 LTGRDFSHDD 2517 TLDVPTQVEL2527 LIKQATSHEN2537 LCQCYIGWCP2547 FW
|
|||||
|
|
||||||
| Click to View More Binding Site Information of This Target and Ligand Pair | ||||||
| Click to View More Binding Site Information of This Target with Different Ligands | ||||||
| Different Human System Profiles of Target | Top |
|---|---|
|
Human Similarity Proteins
of target is determined by comparing the sequence similarity of all human proteins with the target based on BLAST. The similarity proteins for a target are defined as the proteins with E-value < 0.005 and outside the protein families of the target.
A target that has fewer human similarity proteins outside its family is commonly regarded to possess a greater capacity to avoid undesired interactions and thus increase the possibility of finding successful drugs
(Brief Bioinform, 21: 649-662, 2020).
Human Tissue Distribution
of target is determined from a proteomics study that quantified more than 12,000 genes across 32 normal human tissues. Tissue Specificity (TS) score was used to define the enrichment of target across tissues.
The distribution of targets among different tissues or organs need to be taken into consideration when assessing the target druggability, as it is generally accepted that the wider the target distribution, the greater the concern over potential adverse effects
(Nat Rev Drug Discov, 20: 64-81, 2021).
Human Pathway Affiliation
of target is determined by the life-essential pathways provided on KEGG database. The target-affiliated pathways were defined based on the following two criteria (a) the pathways of the studied target should be life-essential for both healthy individuals and patients, and (b) the studied target should occupy an upstream position in the pathways and therefore had the ability to regulate biological function.
Targets involved in a fewer pathways have greater likelihood to be successfully developed, while those associated with more human pathways increase the chance of undesirable interferences with other human processes
(Pharmacol Rev, 58: 259-279, 2006).
Biological Network Descriptors
of target is determined based on a human protein-protein interactions (PPI) network consisting of 9,309 proteins and 52,713 PPIs, which were with a high confidence score of ≥ 0.95 collected from STRING database.
The network properties of targets based on protein-protein interactions (PPIs) have been widely adopted for the assessment of target’s druggability. Proteins with high node degree tend to have a high impact on network function through multiple interactions, while proteins with high betweenness centrality are regarded to be central for communication in interaction networks and regulate the flow of signaling information
(Front Pharmacol, 9, 1245, 2018;
Curr Opin Struct Biol. 44:134-142, 2017).
Human Similarity Proteins
Human Tissue Distribution
Human Pathway Affiliation
Biological Network Descriptors
|
|
|
There is no similarity protein (E value < 0.005) for this target
|
|
Note:
If a protein has TS (tissue specficity) scores at least in one tissue >= 2.5, this protein is called tissue-enriched (including tissue-enriched-but-not-specific and tissue-specific). In the plots, the vertical lines are at thresholds 2.5 and 4.
|
| KEGG Pathway | Pathway ID | Affiliated Target | Pathway Map |
|---|---|---|---|
| ErbB signaling pathway | hsa04012 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| HIF-1 signaling pathway | hsa04066 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Phospholipase D signaling pathway | hsa04072 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Autophagy - other | hsa04136 | Affiliated Target |
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| Autophagy - animal | hsa04140 | Affiliated Target |
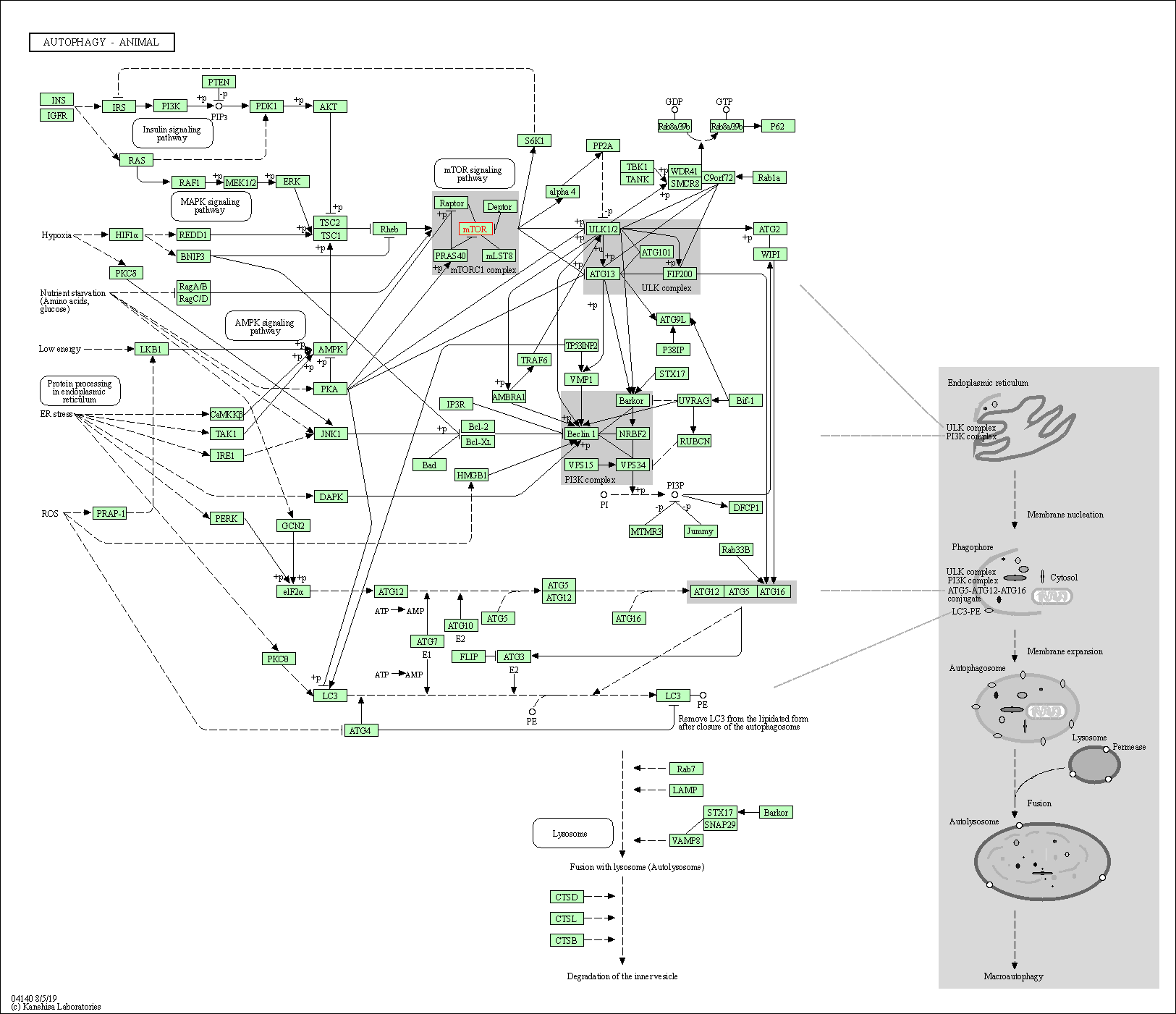
|
| Class: Cellular Processes => Transport and catabolism | Pathway Hierarchy | ||
| mTOR signaling pathway | hsa04150 | Affiliated Target |
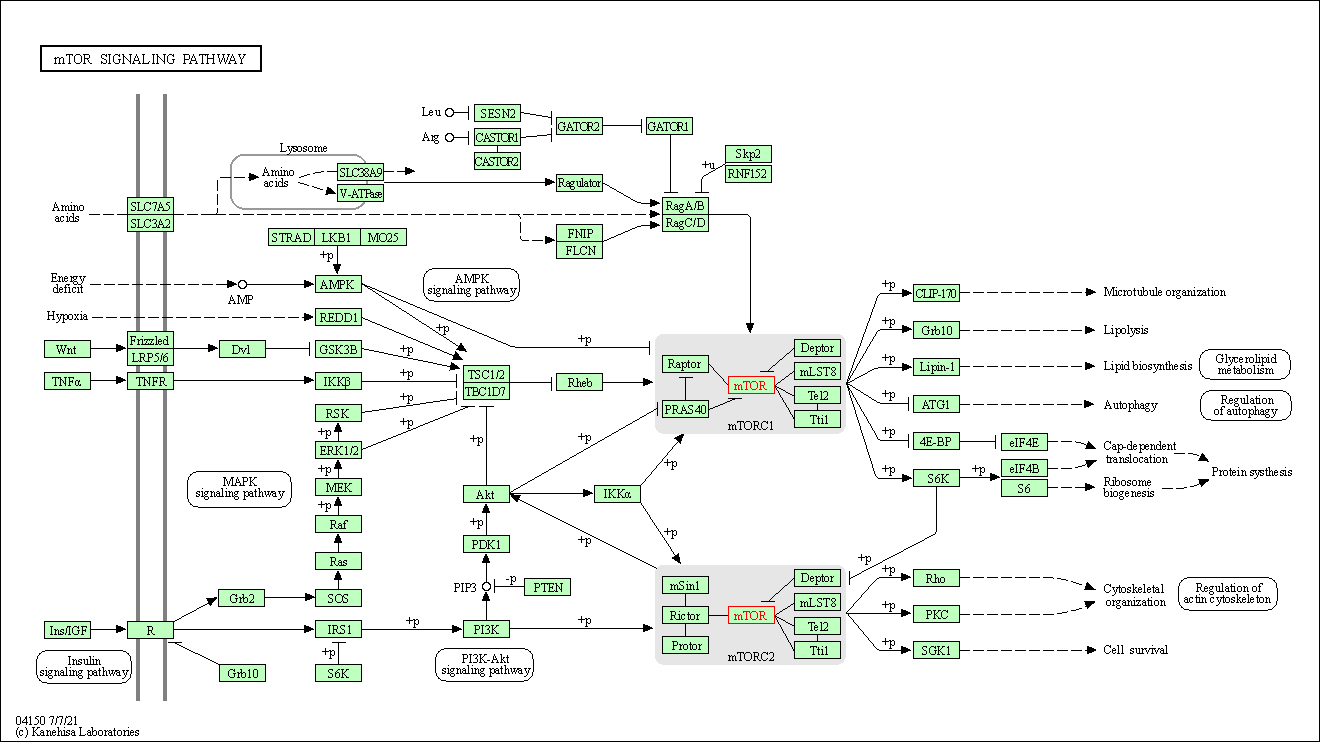
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| PI3K-Akt signaling pathway | hsa04151 | Affiliated Target |
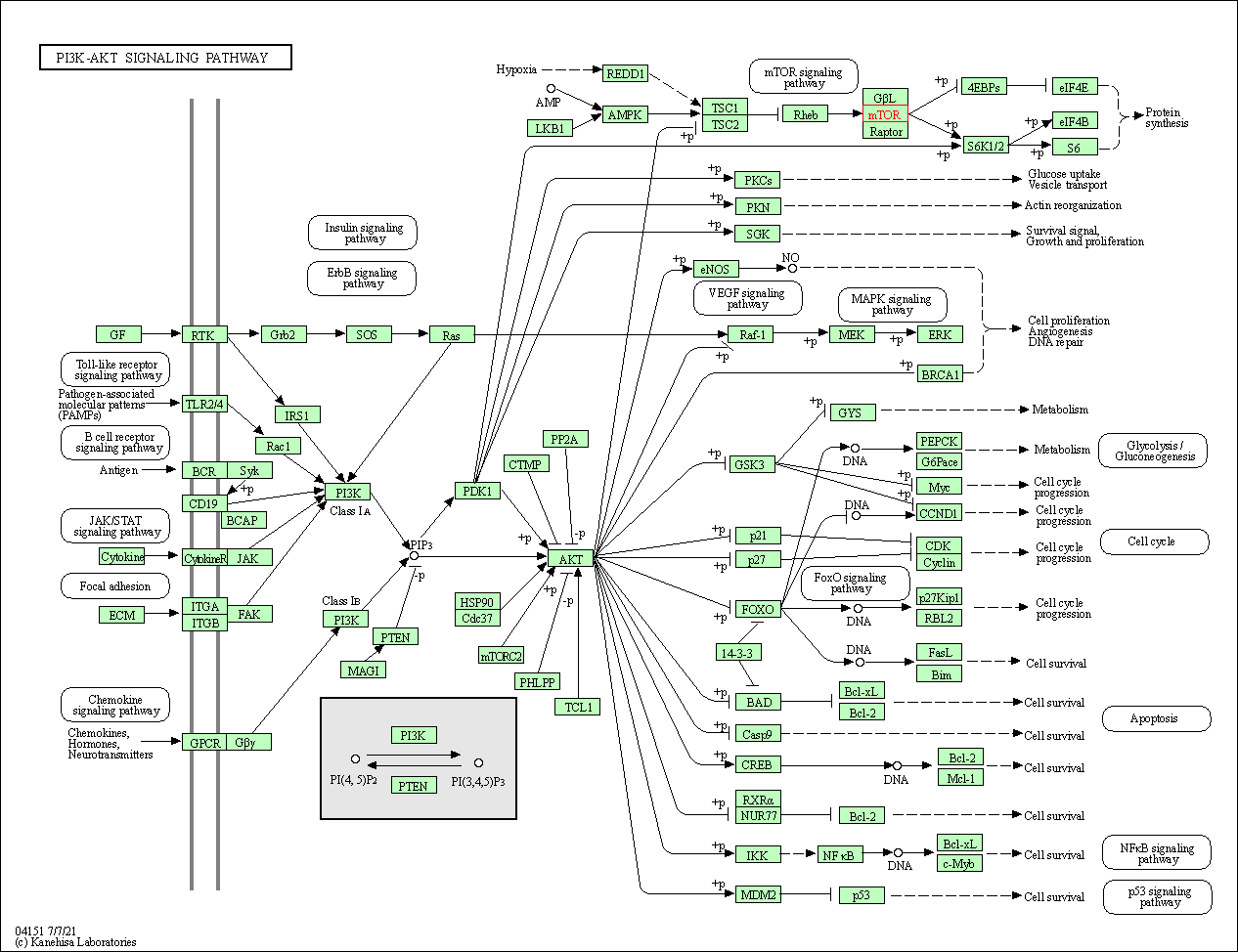
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| AMPK signaling pathway | hsa04152 | Affiliated Target |
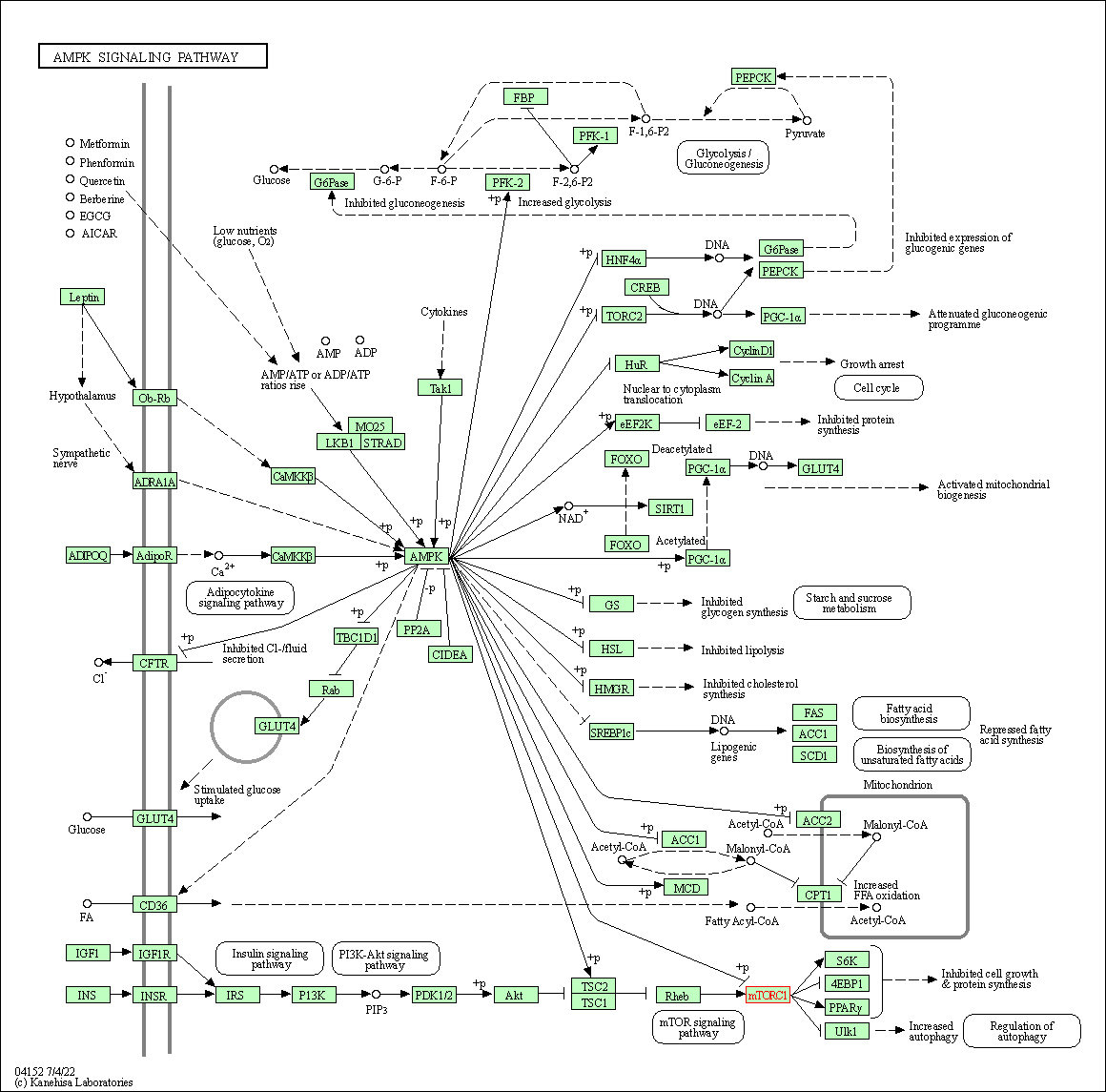
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Longevity regulating pathway | hsa04211 | Affiliated Target |
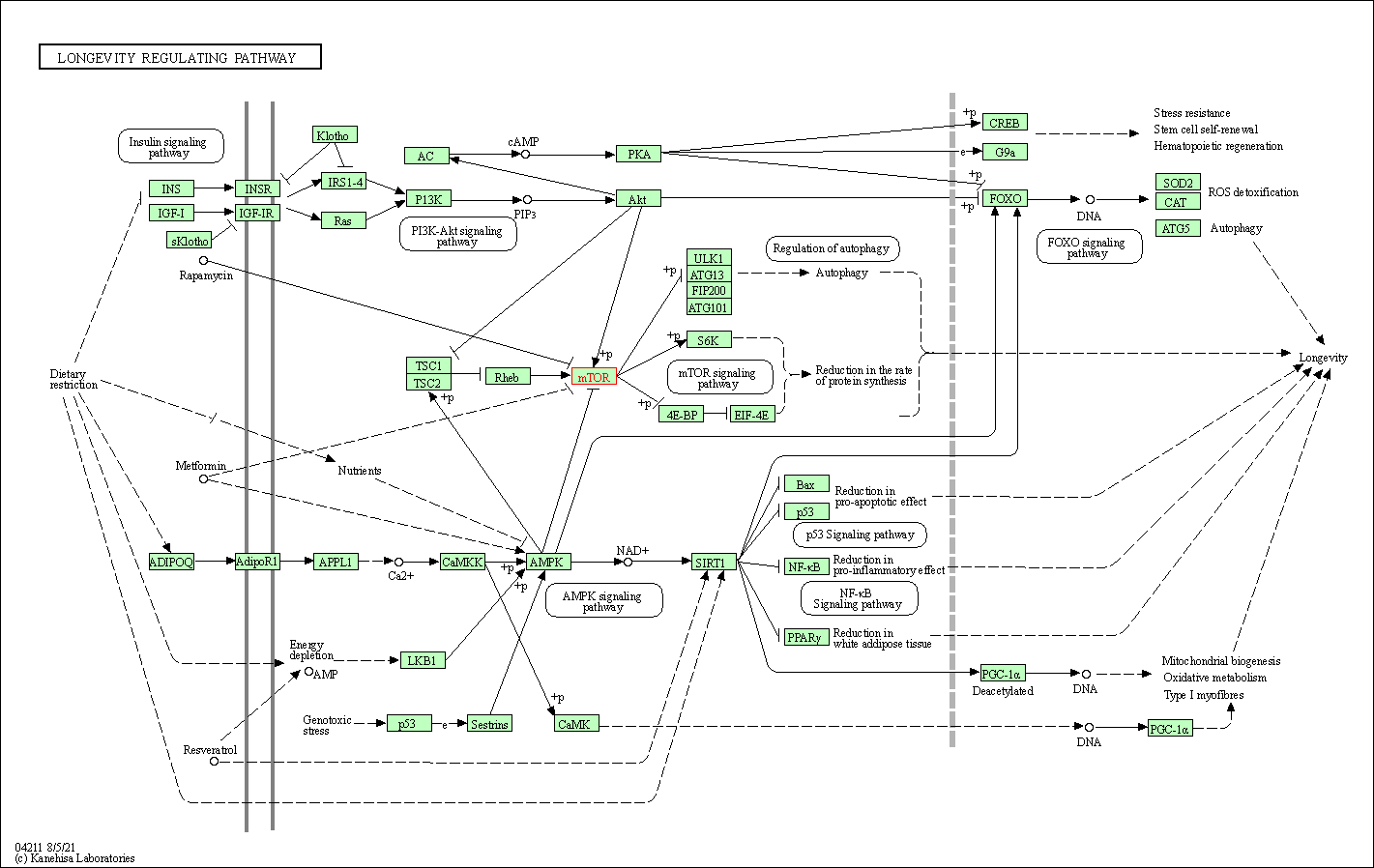
|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Longevity regulating pathway - multiple species | hsa04213 | Affiliated Target |
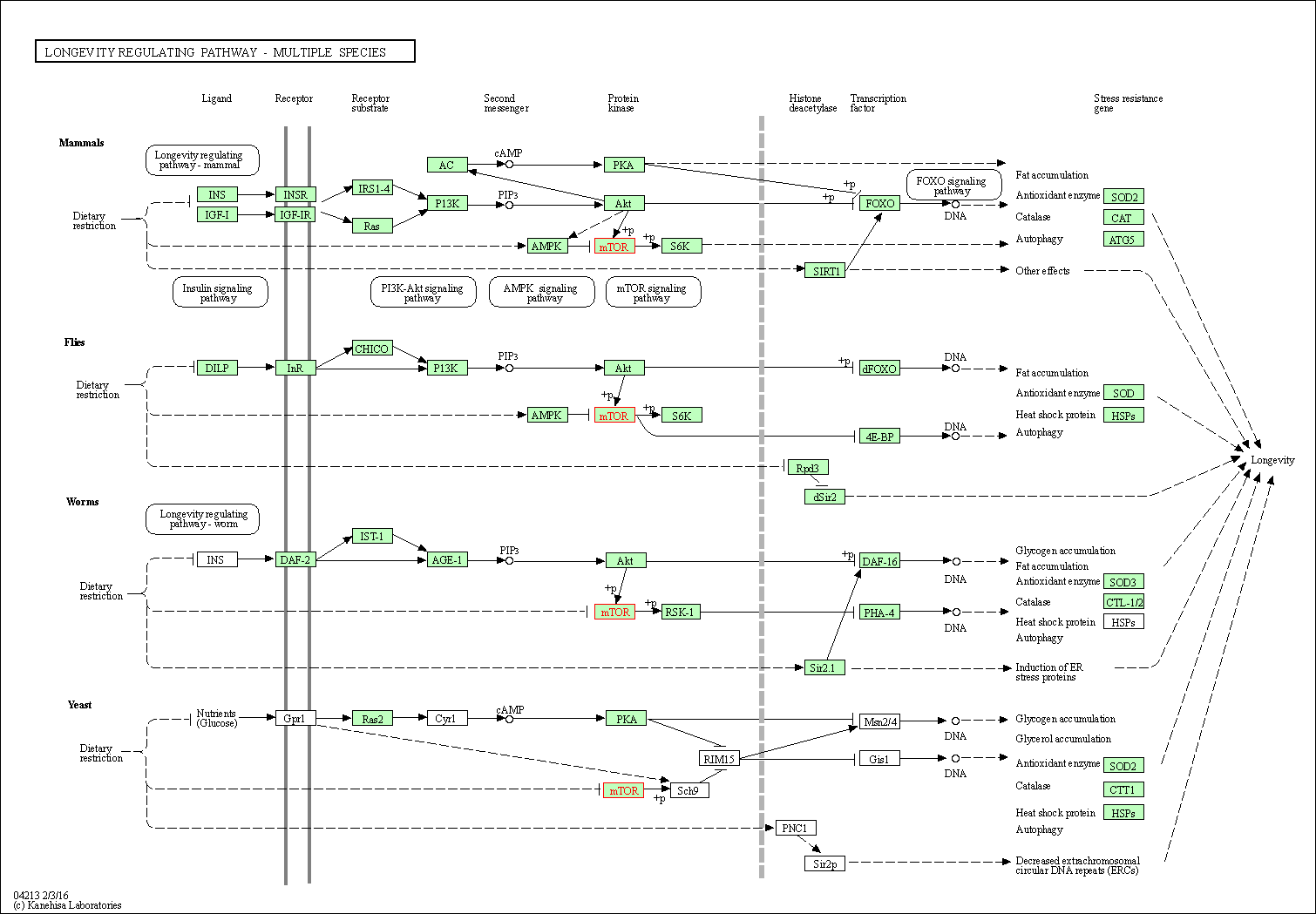
|
| Class: Organismal Systems => Aging | Pathway Hierarchy | ||
| Cellular senescence | hsa04218 | Affiliated Target |
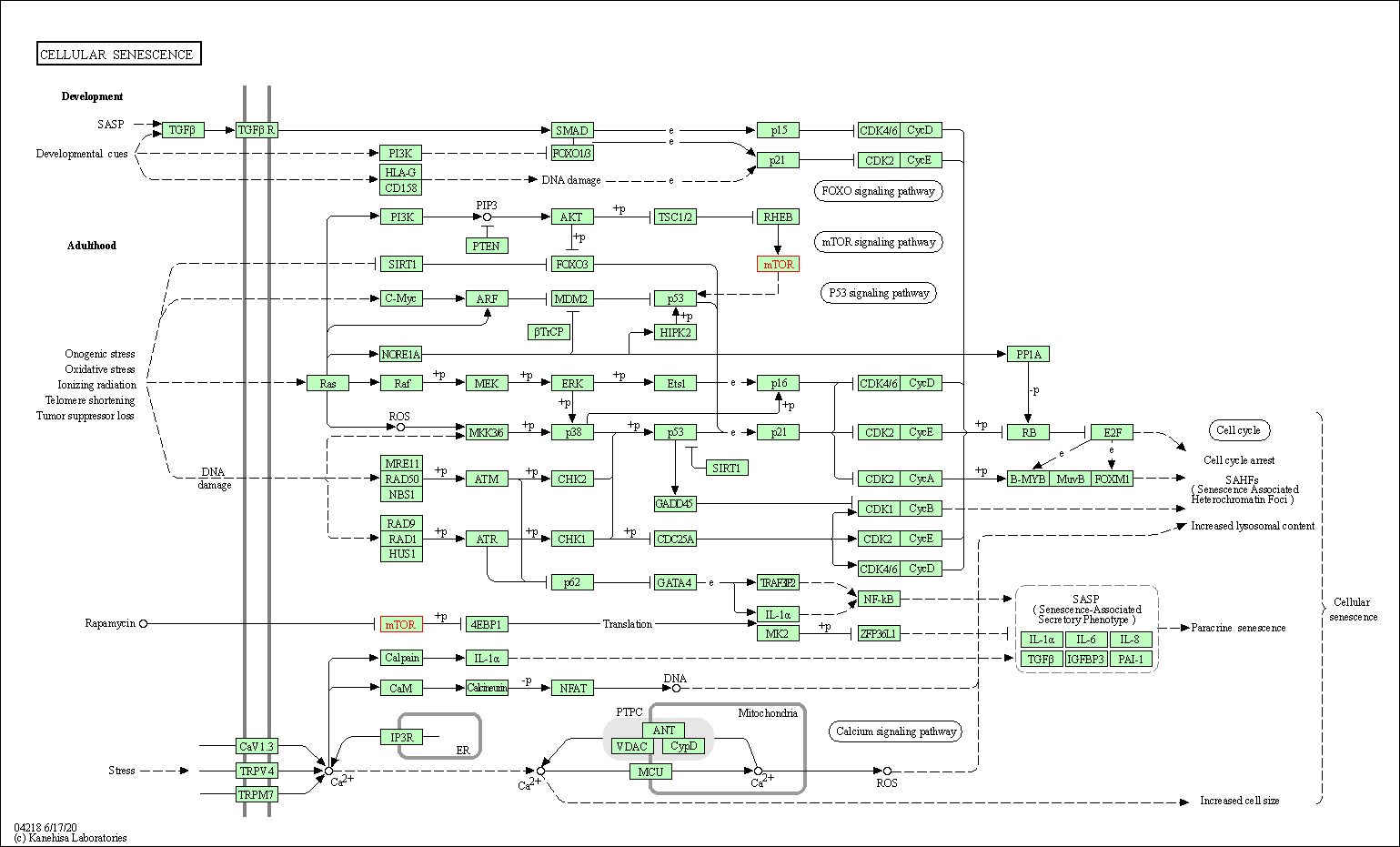
|
| Class: Cellular Processes => Cell growth and death | Pathway Hierarchy | ||
| Apelin signaling pathway | hsa04371 | Affiliated Target |
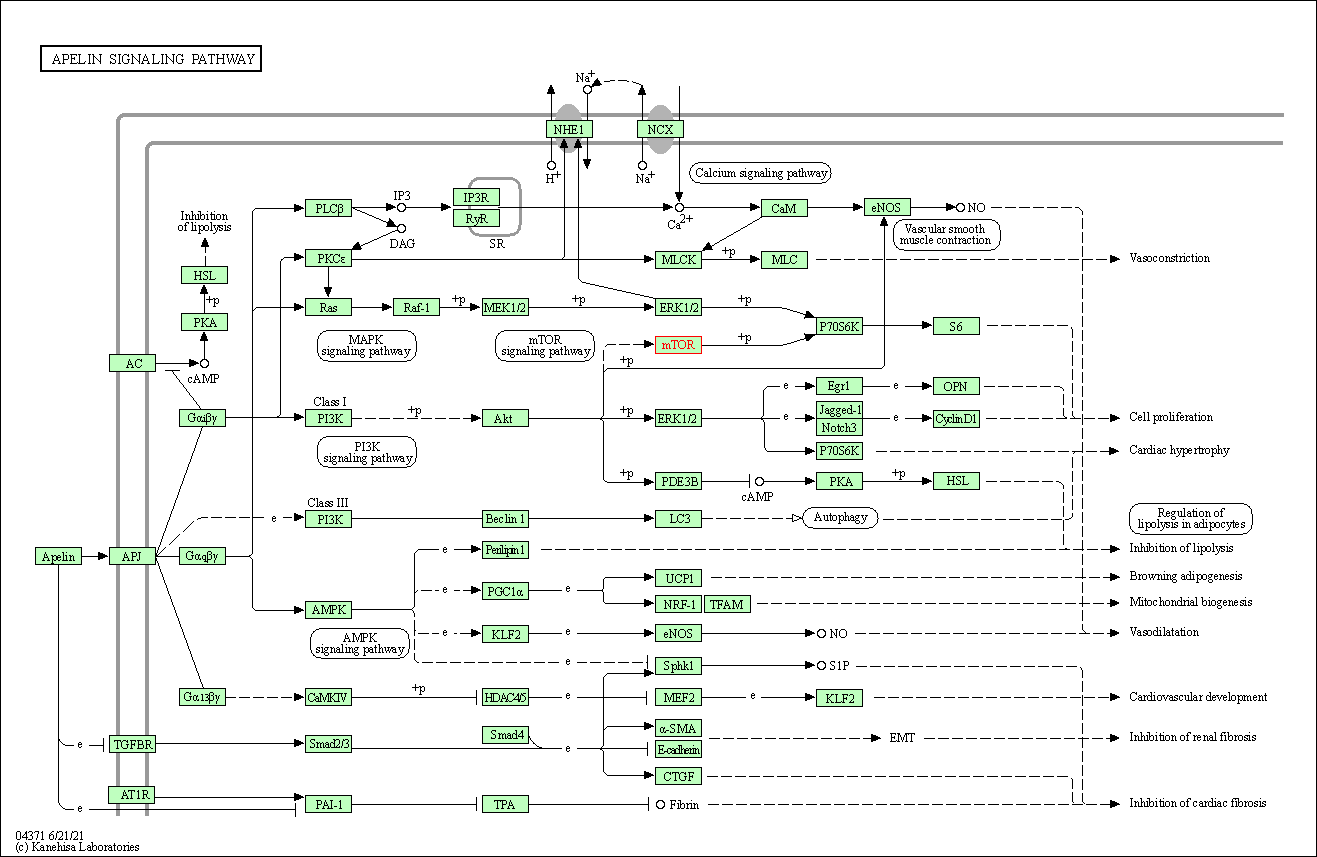
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Neutrophil extracellular trap formation | hsa04613 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| JAK-STAT signaling pathway | hsa04630 | Affiliated Target |
|
| Class: Environmental Information Processing => Signal transduction | Pathway Hierarchy | ||
| Th17 cell differentiation | hsa04659 | Affiliated Target |
|
| Class: Organismal Systems => Immune system | Pathway Hierarchy | ||
| Thermogenesis | hsa04714 | Affiliated Target |
|
| Class: Organismal Systems => Environmental adaptation | Pathway Hierarchy | ||
| Insulin signaling pathway | hsa04910 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Thyroid hormone signaling pathway | hsa04919 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Adipocytokine signaling pathway | hsa04920 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Growth hormone synthesis, secretion and action | hsa04935 | Affiliated Target |
|
| Class: Organismal Systems => Endocrine system | Pathway Hierarchy | ||
| Click to Show/Hide the Information of Affiliated Human Pathways | |||
| Degree | 68 | Degree centrality | 7.31E-03 | Betweenness centrality | 1.09E-02 |
|---|---|---|---|---|---|
| Closeness centrality | 2.67E-01 | Radiality | 1.46E+01 | Clustering coefficient | 1.22E-01 |
| Neighborhood connectivity | 3.47E+01 | Topological coefficient | 3.19E-02 | Eccentricity | 11 |
| Download | Click to Download the Full PPI Network of This Target | ||||
| Chemical Structure based Activity Landscape of Target | Top |
|---|---|
| Target Poor or Non Binders | Top | |||||
|---|---|---|---|---|---|---|
| Target Poor or Non Binders | ||||||
| References | Top | |||||
|---|---|---|---|---|---|---|
| REF 1 | Coronaviruses - drug discovery and therapeutic options. Nat Rev Drug Discov. 2016 May;15(5):327-47. | |||||
| REF 2 | Drugs@FDA. U.S. Food and Drug Administration. U.S. Department of Health & Human Services. 2015 | |||||
| REF 3 | FDA Approved Drug Products from FDA Official Website. 2019. Application Number: (ANDA) 201578. | |||||
| REF 4 | ClinicalTrials.gov (NCT02049957) Safety and Efficacy Study of Sapanisertib in Combination With Exemestane or Fulvestrant in Postmenopausal Women With Estrogen Receptor Positive/Human Epidermal Growth Factor Receptor 2 Negative (ER+/HER2-) Metastatic Breast Cancer. U.S. National Institutes of Health. | |||||
| REF 5 | Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020 Mar 16;6:14. | |||||
| REF 6 | Prevent COVID-19 Severity by Repurposing mTOR Inhibitors. 2 April 2020 | |||||
| REF 7 | Large FK506-binding proteins shape the pharmacology of rapamycin. Mol Cell Biol. 2013 Apr;33(7):1357-67. | |||||
| REF 8 | The 3.2-? resolution structure of human mTORC2. Sci Adv. 2020 Nov 6;6(45):eabc1251. | |||||
If You Find Any Error in Data or Bug in Web Service, Please Kindly Report It to Dr. Zhou and Dr. Zhang.

